Genomic investigations of unexplained acute hepatitis in children
- PMID: 36996872
- PMCID: PMC10170458
- DOI: 10.1038/s41586-023-06003-w
Genomic investigations of unexplained acute hepatitis in children
Abstract
Since its first identification in Scotland, over 1,000 cases of unexplained paediatric hepatitis in children have been reported worldwide, including 278 cases in the UK1. Here we report an investigation of 38 cases, 66 age-matched immunocompetent controls and 21 immunocompromised comparator participants, using a combination of genomic, transcriptomic, proteomic and immunohistochemical methods. We detected high levels of adeno-associated virus 2 (AAV2) DNA in the liver, blood, plasma or stool from 27 of 28 cases. We found low levels of adenovirus (HAdV) and human herpesvirus 6B (HHV-6B) in 23 of 31 and 16 of 23, respectively, of the cases tested. By contrast, AAV2 was infrequently detected and at low titre in the blood or the liver from control children with HAdV, even when profoundly immunosuppressed. AAV2, HAdV and HHV-6 phylogeny excluded the emergence of novel strains in cases. Histological analyses of explanted livers showed enrichment for T cells and B lineage cells. Proteomic comparison of liver tissue from cases and healthy controls identified increased expression of HLA class 2, immunoglobulin variable regions and complement proteins. HAdV and AAV2 proteins were not detected in the livers. Instead, we identified AAV2 DNA complexes reflecting both HAdV-mediated and HHV-6B-mediated replication. We hypothesize that high levels of abnormal AAV2 replication products aided by HAdV and, in severe cases, HHV-6B may have triggered immune-mediated hepatic disease in genetically and immunologically predisposed children.
© 2023. The Author(s).
Conflict of interest statement
.J.B. is a member of the MHRA COVID Vaccines and Therapeutics committees; holder of Wellcome Trust, UKRI and NIHR funding; and principal investigator on the GSK LUNAR study to provide MHRA with data on SARS-CoV-2 sequences in patients treated with sotrovimab.
Figures
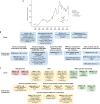
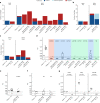

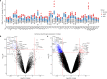
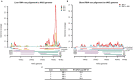


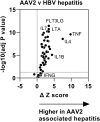
Comment in
-
Severe hepatitis outbreak in children linked to AAV2 virus.Nature. 2023 May;617(7961):471-472. doi: 10.1038/d41586-023-00570-8. Nature. 2023. PMID: 36997704 No abstract available.
References
-
- ECDC–WHO. Joint ECDC–WHO Regional Office for Europe hepatitis of unknown origin in children surveillance bulletin. ECDChttps://cdn.ecdc.europa.eu/novhep-surveillance/ (2022).
-
- UKHSA. Investigation into acute hepatitis of unknown aetiology in children in England: technical briefing 3 (UKHSA, 2022).
-
- UKHSA. Investigation into acute hepatitis of unknown aetiology in children in England: technical briefing 4 (UKHSA, 2022).
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

