Adeno-associated virus 2 infection in children with non-A-E hepatitis
- PMID: 36996873
- PMCID: PMC7617659
- DOI: 10.1038/s41586-023-05948-2
Adeno-associated virus 2 infection in children with non-A-E hepatitis
Abstract
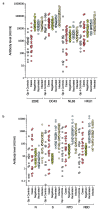
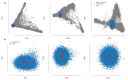
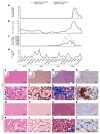
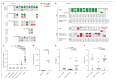
An outbreak of acute hepatitis of unknown aetiology in children was reported in Scotland1 in April 2022 and has now been identified in 35 countries2. Several recent studies have suggested an association with human adenovirus with this outbreak, a virus not commonly associated with hepatitis. Here we report a detailed case-control investigation and find an association between adeno-associated virus 2 (AAV2) infection and host genetics in disease susceptibility. Using next-generation sequencing, PCR with reverse transcription, serology and in situ hybridization, we detected recent infection with AAV2 in plasma and liver samples in 26 out of 32 (81%) cases of hepatitis compared with 5 out of 74 (7%) of samples from unaffected individuals. Furthermore, AAV2 was detected within ballooned hepatocytes alongside a prominent T cell infiltrate in liver biopsy samples. In keeping with a CD4+ T-cell-mediated immune pathology, the human leukocyte antigen (HLA) class II HLA-DRB1*04:01 allele was identified in 25 out of 27 cases (93%) compared with a background frequency of 10 out of 64 (16%; P = 5.49 × 10-12). In summary, we report an outbreak of acute paediatric hepatitis associated with AAV2 infection (most likely acquired as a co-infection with human adenovirus that is usually required as a 'helper virus' to support AAV2 replication) and disease susceptibility related to HLA class II status.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures




Comment in
-
Severe hepatitis outbreak in children linked to AAV2 virus.Nature. 2023 May;617(7961):471-472. doi: 10.1038/d41586-023-00570-8. Nature. 2023. PMID: 36997704 No abstract available.
References
-
- Severe acute hepatitis of unknown aetiology in children—multi-country. World Health Organization; 2022. Jul 12, https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON400 .
-
- Investigation into Acute Hepatitis of Unknown Aetiology in Children in England Technical Briefing 4. UK Health Security Agency; 2022.
-
- Karpen SJ. Acute hepatitis in children in 2022—human adenovirus 41? N Engl J Med. 2022 - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- MC_UU_00007/10/MRC_/Medical Research Council/United Kingdom
- R01 AI158861/AI/NIAID NIH HHS/United States
- MC_UU_12014/1/MRC_/Medical Research Council/United Kingdom
- 223164/WT_/Wellcome Trust/United Kingdom
- MR/W005611/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00034/6/MRC_/Medical Research Council/United Kingdom
- MC_PC_19025/MRC_/Medical Research Council/United Kingdom
- MC_UU_00034/5/MRC_/Medical Research Council/United Kingdom
- MC_PC_20029/MRC_/Medical Research Council/United Kingdom
- MC_PC_19059/MRC_/Medical Research Council/United Kingdom
- CO-CIN-01/DH_/Department of Health/United Kingdom
- 206369/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- 206369/WT_/Wellcome Trust/United Kingdom
- MR/W030454/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_22004/MRC_/Medical Research Council/United Kingdom
- MR/X010252/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12014/12/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

