UPF3A is dispensable for nonsense-mediated mRNA decay in mouse pluripotent and somatic cells
- PMID: 36997282
- PMCID: PMC10070813
- DOI: 10.26508/lsa.202201589
UPF3A is dispensable for nonsense-mediated mRNA decay in mouse pluripotent and somatic cells
Abstract
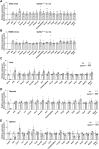
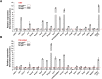
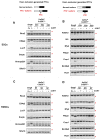
Nonsense-mediated mRNA decay (NMD) is a highly conserved regulatory mechanism of post-transcriptional gene expression in eukaryotic cells. NMD plays essential roles in mRNA quality and quantity control and thus safeguards multiple biological processes including embryonic stem cell differentiation and organogenesis. UPF3A and UPF3B in vertebrate species, originated from a single UPF3 gene in yeast, are key factors in the NMD machinery. Although UPF3B is a well-recognized weak NMD-promoting factor, whether UPF3A functions in promoting or suppressing NMD is under debate. In this study, we generated a Upf3a conditional knockout mouse strain and established multiple lines of embryonic stem cells and somatic cells without UPF3A. Through extensive analysis on the expressions of 33 NMD targets, we found UPF3A neither represses NMD in mouse embryonic stem cells, somatic cells, nor in major organs including the liver, spleen, and thymus. Our study reinforces that UPF3A is dispensable for NMD when UPF3B is present. Furthermore, UPF3A may weakly and selectively promote NMD in certain murine organs.
© 2023 Chen et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







References
-
- Bufton JC, Powers KT, Szeto JYA, Toelzer C, Berger I, Schaffitzel C (2022) Structures of nonsense-mediated mRNA decay factors upf3b and upf3a in complex with upf2 reveal molecular basis for competitive binding and for neurodevelopmental disorder-causing mutation. Nucleic Acids Res 50: 5934–5947. 10.1093/nar/gkac421 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
