A Vaccinia-based system for directed evolution of GPCRs in mammalian cells
- PMID: 36997531
- PMCID: PMC10063554
- DOI: 10.1038/s41467-023-37191-8
A Vaccinia-based system for directed evolution of GPCRs in mammalian cells
Abstract
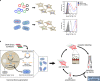
Directed evolution in bacterial or yeast display systems has been successfully used to improve stability and expression of G protein-coupled receptors for structural and biophysical studies. Yet, several receptors cannot be tackled in microbial systems due to their complex molecular composition or unfavorable ligand properties. Here, we report an approach to evolve G protein-coupled receptors in mammalian cells. To achieve clonality and uniform expression, we develop a viral transduction system based on Vaccinia virus. By rational design of synthetic DNA libraries, we first evolve neurotensin receptor 1 for high stability and expression. Second, we demonstrate that receptors with complex molecular architectures and large ligands, such as the parathyroid hormone 1 receptor, can be readily evolved. Importantly, functional receptor properties can now be evolved in the presence of the mammalian signaling environment, resulting in receptor variants exhibiting increased allosteric coupling between the ligand binding site and the G protein interface. Our approach thus provides insights into the intricate molecular interplay required for GPCR activation.
© 2023. The Author(s).
Conflict of interest statement
M.S., S.S., L.M., E.G., M.Z. and E.S.S. are employees of Vaccinex, Inc. and own stock and/or stock options in the company. R.S. is an employee of MorphoSys AG and declares that no competing interests exist. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

