Bursts in biosynthetic gene cluster transcription are accompanied by surges of natural compound production in the myxobacterium Sorangium sp
- PMID: 36998231
- PMCID: PMC10128143
- DOI: 10.1111/1751-7915.14246
Bursts in biosynthetic gene cluster transcription are accompanied by surges of natural compound production in the myxobacterium Sorangium sp
Abstract
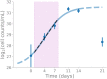
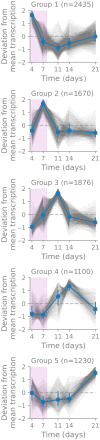
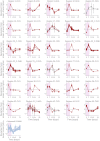
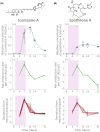
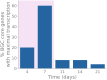
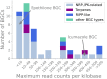
A better understanding of the genetic regulation of the biosynthesis of microbial compounds could accelerate the discovery of new biologically active molecules and facilitate their production. To this end, we have investigated the time course of genome-wide transcription in the myxobacterium Sorangium sp. So ce836 in relation to its production of natural compounds. Time-resolved RNA sequencing revealed that core biosynthesis genes from 48 biosynthetic gene clusters (BGCs; 92% of all BGCs encoded in the genome) were actively transcribed at specific time points in a batch culture. The majority (80%) of polyketide synthase and non-ribosomal peptide synthetase genes displayed distinct peaks of transcription during exponential bacterial growth. Strikingly, these bursts in BGC transcriptional activity were associated with surges in the net production rates of known natural compounds, indicating that their biosynthesis was critically regulated at the transcriptional level. In contrast, BGC read counts from single time points had limited predictive value about biosynthetic activity, since transcription levels varied >100-fold among BGCs with detected natural products. Taken together, our time-course data provide unique insights into the dynamics of natural compound biosynthesis and its regulation in a wild-type myxobacterium, challenging the commonly cited notion of preferential BGC expression under nutrient-limited conditions. The close association observed between BGC transcription and compound production warrants additional efforts to develop genetic engineering tools for boosting compound yields from myxobacterial producer strains.
© 2023 The Authors. Microbial Biotechnology published by Applied Microbiology International and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they do not have any competing interests.
Figures
References
-
- Aigle, B. , Lautru, S. , Spiteller, D. , Dickschat, J.S. , Challis, G.L. , Leblond, P. et al. (2014) Genome mining of Streptomyces ambofaciens . Journal of Industrial Microbiology & Biotechnology, 41, 251–263. - PubMed
-
- Altmann, K.‐H. , Höfle, G. , Müller, R. , Mulzer, J. & Prantz, K. (2009) The epothilones: an outstanding family of anti‐tumor agents. Vienna: Springer. - PubMed
-
- Amos, G.C.A. , Awakawa, T. , Tuttle, R.N. , Letzel, A.‐C. , Kim, M.C. , Kudo, Y. et al. (2017) Comparative transcriptomics as a guide to natural product discovery and biosynthetic gene cluster functionality. Proceedings of the National Academy of Sciences of the United States of America, 114, E11121–E11130. - PMC - PubMed
-
- Baltz, R.H. (2017) Gifted microbes for genome mining and natural product discovery. Journal of Industrial Microbiology & Biotechnology, 44, 573–588. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

