Systematic identification of potential key microRNAs and circRNAs in the dorsal root ganglia of mice with sciatic nerve injury
- PMID: 36998510
- PMCID: PMC10043392
- DOI: 10.3389/fnmol.2023.1119164
Systematic identification of potential key microRNAs and circRNAs in the dorsal root ganglia of mice with sciatic nerve injury
Abstract
Background: Neuropathic pain (NeP) is a pathological condition arising from a lesion or disease affecting the somatosensory system. Accumulating evidence has shown that circular RNAs (circRNAs) exert critical functions in neurodegenerative diseases by sponging microRNAs (miRNAs). However, the functions and regulatory mechanisms of circRNAs as competitive endogenous RNAs (ceRNAs) in NeP remain to be determined.
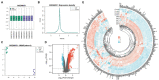
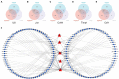
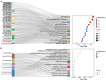
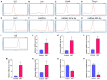
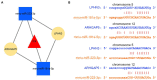
Methods: The sequencing dataset GSE96051 was obtained from the public Gene Expression Omnibus (GEO) database. First, we conducted a comparison of gene expression profiles in the L3/L4 dorsal root ganglion (DRG) of sciatic nerve transection (SNT) mice (n = 5) and uninjured mice (Control) (n = 4) to define the differentially expressed genes (DEGs). Then, critical hub genes were screened by exploring protein-protein interaction (PPI) networks with Cytoscape software, and the miRNAs bound to them were predicted and selected and then validated by qRT-PCR. Furthermore, key circRNAs were predicted and filtered, and the network of circRNA-miRNA-mRNA in NeP was constructed.
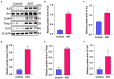
Results: A total of 421 DEGs were identified, including 332 upregulated genes and 89 downregulated genes. Ten hub genes, including IL6, Jun, Cd44, Timp1, and Csf1, were identified. Two miRNAs, mmu-miR-181a-5p and mmu-miR-223-3p, were preliminarily verified as key regulators of NeP development. In addition, circARHGAP5 and circLPHN3 were identified as key circRNAs. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis demonstrated that these differentially expressed mRNAs and targeting miRNAs were involved in signal transduction, positive regulation of receptor-mediated endocytosis and regulation of neuronal synaptic plasticity. These findings have useful implications for the exploration of new mechanisms and therapeutic targets for NeP.
Conclusion: These newly identified miRNAs and circRNAs in networks reveal potential diagnostic or therapeutic targets for NeP.
Keywords: ceRNA; circRNA; dorsal root ganglia; miR-181a-5p; miR-223-3p; microRNA; neuropathic pain.
Copyright © 2023 Yu, Xu, Lin and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

