Epigenetic immune monitoring for COVID-19 disease course prognosis
- PMID: 36999021
- PMCID: PMC10043382
- DOI: 10.3389/fimmu.2023.1107900
Epigenetic immune monitoring for COVID-19 disease course prognosis
Erratum in
-
Corrigendum: Epigenetic immune monitoring for COVID-19 disease course prognosis.Front Immunol. 2024 Jun 27;15:1451571. doi: 10.3389/fimmu.2024.1451571. eCollection 2024. Front Immunol. 2024. PMID: 38994367 Free PMC article.
Abstract
Background: The course of COVID-19 is associated with severe dysbalance of the immune system, causing both leukocytosis and lymphopenia. Immune cell monitoring may be a powerful tool to prognosticate disease outcome. However, SARS-CoV-2 positive subjects are isolated upon initial diagnosis, thus barring standard immune monitoring using fresh blood. This dilemma may be solved by epigenetic immune cell counting.
Methods: In this study, we used epigenetic immune cell counting by qPCR as an alternative way of quantitative immune monitoring for venous blood, capillary blood dried on filter paper (dried blood spots, DBS) and nasopharyngeal swabs, potentially allowing a home-based monitoring approach.
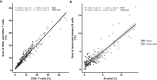
Results: Epigenetic immune cell counting in venous blood showed equivalence with dried blood spots and with flow cytometrically determined cell counts of venous blood in healthy subjects. In venous blood, we detected relative lymphopenia, neutrophilia, and a decreased lymphocyte-to-neutrophil ratio for COVID-19 patients (n =103) when compared with healthy donors (n = 113). Along with reported sex-related differences in survival we observed dramatically lower regulatory T cell counts in male patients. In nasopharyngeal swabs, T and B cell counts were significantly lower in patients compared to healthy subjects, mirroring the lymphopenia in blood. Naïve B cell frequency was lower in severely ill patients than in patients with milder stages.
Conclusions: Overall, the analysis of immune cell counts is a strong predictor of clinical disease course and the use of epigenetic immune cell counting by qPCR may provide a tool that can be used even for home-isolated patients.
Keywords: COVID-19; SARS-CoV-2; disease prognosis; epigenetic qPCR; immune monitoring; lymphopenia.
Copyright © 2023 Samans, Rosselló Chornet, Rosselló Chornet, Jung, Schildknecht, Lozza, Alos Zaragoza, Hernández Laforet, Babel and Olek.
Conflict of interest statement
Authors BS, AR, KS, JJ, LL and SO were employed by the company Precision for Medicine. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Coronavirus disease (COVID-19) . Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
-
- Office of the Assistant Secretary for Preparedness H. Pandemic Influenza Plan - Update IV . (2017).
-
- Mohan SS, McDermott BP, Cunha BA. The diagnostic and prognostic significance of relative lymphopenia in adult patients with influenza a. Am J Med (2005) 118(11):1307. doi: 10.1016/j.amjmed.2005.06.018 - DOI
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

