RNA origami scaffolds facilitate cryo-EM characterization of a Broccoli-Pepper aptamer FRET pair
- PMID: 36999628
- PMCID: PMC10201433
- DOI: 10.1093/nar/gkad224
RNA origami scaffolds facilitate cryo-EM characterization of a Broccoli-Pepper aptamer FRET pair
Abstract
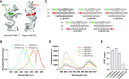
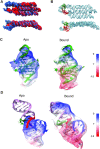
Cryogenic electron microscopy (cryo-EM) is a promising method for characterizing the structure of larger RNA structures and complexes. However, the structure of individual aptamers is difficult to solve by cryo-EM due to their low molecular weight and a high signal-to-noise ratio. By placing RNA aptamers on larger RNA scaffolds, the contrast for cryo-EM can be increased to allow the determination of the tertiary structure of the aptamer. Here we use the RNA origami method to scaffold two fluorescent aptamers (Broccoli and Pepper) in close proximity and show that their cognate fluorophores serve as donor and acceptor for FRET. Next, we use cryo-EM to characterize the structure of the RNA origami with the two aptamers to a resolution of 4.4 Å. By characterizing the aptamers with and without ligand, we identify two distinct modes of ligand binding, which are further supported by selective chemical probing. 3D variability analysis of the cryo-EM data show that the relative position between the two bound fluorophores on the origami fluctuate by only 3.5 Å. Our results demonstrate a general approach for using RNA origami scaffolds for characterizing small RNA motifs by cryo-EM and for positioning functional RNA motifs with high spatial precision.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Jiang F., Kumar R.A., Jones R.A., Patel D.J.. Structural basis of RNA folding and recognition in an AMP-RNA aptamer complex. Nature. 1996; 382:183–186. - PubMed
-
- Nahvi A., Sudarsan N., Ebert M.S., Zou X., Brown K.L., Breaker R.R.. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002; 9:1043. - PubMed
-
- Kruger K., Grabowski P.J., Zaug A.J., Sands J., Gottschling D.E., Cech T.R.. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell. 1982; 31:147–157. - PubMed
-
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S.. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983; 35:849–857. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

