C-terminal frameshift variant of TDP-43 with pronounced aggregation-propensity causes rimmed vacuole myopathy but not ALS/FTD
- PMID: 37000196
- PMCID: PMC10175433
- DOI: 10.1007/s00401-023-02565-1
C-terminal frameshift variant of TDP-43 with pronounced aggregation-propensity causes rimmed vacuole myopathy but not ALS/FTD
Abstract
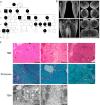
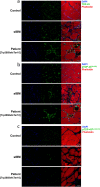
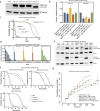
Neuronal TDP-43-positive inclusions are neuropathological hallmark lesions in frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS). Pathogenic missense variants in TARDBP, the gene encoding TDP-43, can cause ALS and cluster in the C-terminal prion-like domain (PrLD), where they modulate the liquid condensation and aggregation properties of the protein. TDP-43-positive inclusions are also found in rimmed vacuole myopathies, including sporadic inclusion body myositis, but myopathy-causing TDP-43 variants have not been reported. Using genome-wide linkage analysis and whole exome sequencing in an extended five-generation family with an autosomal dominant rimmed vacuole myopathy, we identified a conclusively linked frameshift mutation in TDP-43 producing a C-terminally altered PrLD (TDP-43p.Trp385IlefsTer10) (maximum multipoint LOD-score 3.61). Patient-derived muscle biopsies showed TDP-43-positive sarcoplasmic inclusions, accumulation of autophagosomes and transcriptomes with abnormally spliced sarcomeric genes (including TTN and NEB) and increased expression of muscle regeneration genes. In vitro phase separation assays demonstrated that TDP-43Trp385IlefsTer10 does not form liquid-like condensates and readily forms solid-like fibrils indicating increased aggregation propensity compared to wild-type TDP-43. In Drosophila TDP-43p.Trp385IlefsTer10 behaved as a partial loss-of-function allele as it was able to rescue the TBPH (fly ortholog of TARDBP) neurodevelopmental lethal null phenotype while showing strongly reduced toxic gain-of-function properties upon overexpression. Accordingly, TDP-43p.Trp385IlefsTer10 showed reduced toxicity in a primary rat neuron disease model. Together, these genetic, pathological, in vitro and in vivo results demonstrate that TDP-43p.Trp385IlefsTer10 is an aggregation-prone partial loss-of-function variant that causes autosomal dominant vacuolar myopathy but not ALS/FTD. Our study genetically links TDP-43 proteinopathy to myodegeneration, and reveals a tissue-specific role of the PrLD in directing pathology.
Keywords: ALS/FTD; Drosophila; Genetics; Myopathy; Phase separation; TDP-43.
© 2023. The Author(s).
Conflict of interest statement
J.S. is a consultant for Dewpoint Therapeutics, Confluence Therapeutics, ADRx, and Neumora. The other authors have nothing to declare.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

