Quantitative determination of SLC2A1 variant functional effects in GLUT1 deficiency syndrome
- PMID: 37000947
- PMCID: PMC10187726
- DOI: 10.1002/acn3.51767
Quantitative determination of SLC2A1 variant functional effects in GLUT1 deficiency syndrome
Abstract
Objective: The goal of this study is to demonstrate the utility of a growth assay to quantify the functional impact of single nucleotide variants (SNVs) in SLC2A1, the gene responsible for Glut1DS.
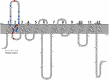
Methods: The functional impact of 40 SNVs in SLC2A1 was quantitatively determined in HAP1 cells in which SLC2A1 is required for growth. Donor libraries were introduced into the endogenous SLC2A1 gene in HAP1-Lig4KO cells using CRISPR/Cas9. Cell populations were harvested and sequenced to quantify the effect of variants on growth and generate a functional score. Quantitative functional scores were compared to 3-OMG uptake, SLC2A1 cell surface expression, CADD score, and clinical data, including CSF/blood glucose ratio.
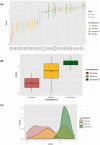
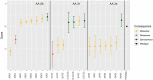
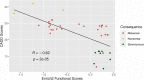
Results: Nonsense variants (N = 3) were reduced in cell culture over time resulting in negative scores (mean score: -1.15 ± 0.17), whereas synonymous variants (N = 10) were not depleted (mean score: 0.25 ± 0.12) (P < 2e-16). Missense variants (N = 27) yielded a range of functional scores including slightly negative scores, supporting a partial function and intermediate phenotype. Several variants with normal results on either cell surface expression (p.N34S and p.W65R) or 3-OMG uptake (p.W65R) had negative functional scores. There is a moderate but significant correlation between our functional scores and CADD scores.
Interpretation: Cell growth is useful to quantitatively determine the functional effects of SLC2A1 variants. Nonsense variants were reliably distinguished from benign variants in this in vitro functional assay. For facilitating early diagnosis and therapeutic intervention, future work is needed to determine the functional effect of every possible variant in SLC2A1.
© 2023 The Authors. Annals of Clinical and Translational Neurology published by Wiley Periodicals LLC on behalf of American Neurological Association.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
