3'-End Processing of Eukaryotic mRNA: Machinery, Regulation, and Impact on Gene Expression
- PMID: 37001138
- PMCID: PMC7614891
- DOI: 10.1146/annurev-biochem-052521-012445
3'-End Processing of Eukaryotic mRNA: Machinery, Regulation, and Impact on Gene Expression
Abstract
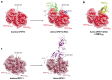
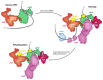
Formation of the 3' end of a eukaryotic mRNA is a key step in the production of a mature transcript. This process is mediated by a number of protein factors that cleave the pre-mRNA, add a poly(A) tail, and regulate transcription by protein dephosphorylation. Cleavage and polyadenylation specificity factor (CPSF) in humans, or cleavage and polyadenylation factor (CPF) in yeast, coordinates these enzymatic activities with each other, with RNA recognition, and with transcription. The site of pre-mRNA cleavage can strongly influence the translation, stability, and localization of the mRNA. Hence, cleavage site selection is highly regulated. The length of the poly(A) tail is also controlled to ensure that every transcript has a similar tail when it is exported from the nucleus. In this review, we summarize new mechanistic insights into mRNA 3'-end processing obtained through structural studies and biochemical reconstitution and outline outstanding questions in the field.
Keywords: endonuclease; poly(A) tail; polyadenylation; polymerase; pre-mRNA processing; transcription.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

