T cell immunotherapies engage neutrophils to eliminate tumor antigen escape variants
- PMID: 37001503
- PMCID: PMC10994488
- DOI: 10.1016/j.cell.2023.03.007
T cell immunotherapies engage neutrophils to eliminate tumor antigen escape variants
Abstract
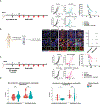
Cancer immunotherapies, including adoptive T cell transfer, can be ineffective because tumors evolve to display antigen-loss-variant clones. Therapies that activate multiple branches of the immune system may eliminate escape variants. Here, we show that melanoma-specific CD4+ T cell therapy in combination with OX40 co-stimulation or CTLA-4 blockade can eradicate melanomas containing antigen escape variants. As expected, early on-target recognition of melanoma antigens by tumor-specific CD4+ T cells was required. Surprisingly, complete tumor eradication was dependent on neutrophils and partly dependent on inducible nitric oxide synthase. In support of these findings, extensive neutrophil activation was observed in mouse tumors and in biopsies of melanoma patients treated with immune checkpoint blockade. Transcriptomic and flow cytometry analyses revealed a distinct anti-tumorigenic neutrophil subset present in treated mice. Our findings uncover an interplay between T cells mediating the initial anti-tumor immune response and neutrophils mediating the destruction of tumor antigen loss variants.
Keywords: CTLA-4; OX40; adoptive T cell therapies; anti-tumor neutrophils; antigenic heterogeneity; immune checkpoint blockade; immunotherapy; neutrophil extracellular traps; neutrophils; tumor hetergeneity.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests T.M. is a consultant for Daiichi Sankyo, Leap Therapeutics, Immunos Therapeutics, and Pfizer and co-founder of Imvaq Therapeutics. T.M. has equity in Imvaq Therapeutics. T.M. reports grants from Bristol Myers Squibb, Surface Oncology, Kyn Therapeutics, Infinity Pharmaceuticals, Peregrine Pharmaceuticals, Adaptive Biotechnologies, Leap Therapeutics, and Aprea. T.M. is an inventor on patent applications related to work on oncolytic viral therapy, alpha virus-based vaccine, neoantigen modeling, CD40, GITR, OX40, PD-1, and CTLA-4. D.H. and S.B. have received royalties from Agenus. R.Z. is an inventor on patent applications related to work on GITR, PD-1, and CTLA-4; has received grant support from Bristol-Myers Squibb; and is a consultant for Leap Therapeutics. J.D.W. is a consultant for Apricity, Ascentage Pharma, AstraZeneca, Bicara Therapeutics, Boehringer Ingelheim, Bristol Myers Squibb, Chugai, Daiichi Sankyo, Dragonfly, Georgiamune, Imvaq, Larkspur, Psioxus, Recepta, Tizona, and Sellas. J.D.W. received grant/research support from Bristol Myers Squibb and Sephora. J.D.W. has equity in Apricity, Arsenal IO, Ascentage, Beigene, CellCarta, Imvaq, Linneaus, Larkspur, Georgiamune, Maverick, Tizona Therapeutics, and Xenimmune. J.D.W. is an inventor on the following patents: xenogeneic DNA vaccines, alphavirus replicon particles expressing TRP2, myeloid-derived suppressor cell (MDSC) assay, Newcastle disease virus for cancer therapy, genomic signature to identify responders to ipilimumab in melanoma, engineered vaccinia viruses for cancer immunotherapy (with T.M.), anti-CD40 agonist mAb fused to monophosphoryl lipid A (MPL) for cancer therapy (with T.M.), CAR(+) T cells targeting differentiation antigens as a means to treat cancer, anti-PD-1 antibody, anti-CTLA4 antibodies, anti-GITR antibodies, and methods of use thereof. D.H. and T.M. are co-inventors on patent applications related to OX40 antibodies. M.E. is a member of the research advisory board for brensocatib for Insmed, Inc.; a member of the scientific advisory board for Vividion Therapeutics, Inc.; and a consultant for Protalix, Inc. and holds shares in Agios Pharmaceuticals, Inc.
Figures
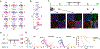

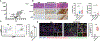
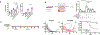


References
-
- Majzner RG, and Mackall CL (2018). Tumor Antigen Escape from CAR T-cell Therapy. Cancer Discov 8, 1219–1226. 10.1158/2159-8290.CD-18-0442. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

