Within-host genetic diversity of SARS-CoV-2 lineages in unvaccinated and vaccinated individuals
- PMID: 37002233
- PMCID: PMC10063955
- DOI: 10.1038/s41467-023-37468-y
Within-host genetic diversity of SARS-CoV-2 lineages in unvaccinated and vaccinated individuals
Abstract
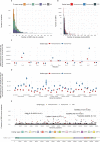
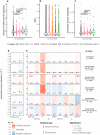
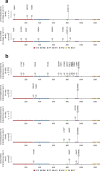
Viral and host factors can shape SARS-CoV-2 evolution. However, little is known about lineage-specific and vaccination-specific mutations that occur within individuals. Here, we analysed deep sequencing data from 2,820 SARS-CoV-2 respiratory samples with different viral lineages to describe the patterns of within-host diversity under different conditions, including vaccine-breakthrough infections. In unvaccinated individuals, variant of Concern (VOC) Alpha, Delta, and Omicron respiratory samples were found to have higher within-host diversity and were under neutral to purifying selection at the full genome level compared to non-VOC SARS-CoV-2. Breakthrough infections in 2-dose or 3-dose Comirnaty and CoronaVac vaccinated individuals did not increase levels of non-synonymous mutations and did not change the direction of selection pressure. Vaccine-induced antibody or T cell responses did not appear to have significant impact on within-host SARS-CoV-2 sequence diversification. Our findings suggest that vaccination does not increase exploration of SARS-CoV-2 protein sequence space and may not facilitate emergence of viral variants.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Update of
-
Within-host diversity of SARS-CoV-2 lineages and effect of vaccination.Res Sq [Preprint]. 2022 Aug 11:rs.3.rs-1927944. doi: 10.21203/rs.3.rs-1927944/v1. Res Sq. 2022. Update in: Nat Commun. 2023 Mar 31;14(1):1793. doi: 10.1038/s41467-023-37468-y. PMID: 35982671 Free PMC article. Updated. Preprint.
References
-
- Mathieu, E. et al. Coronavirus pandemic (COVID-19). Our world in data. Published online at OurWorldInData.org. Retrieved from: https://ourworldindata.org/covid-vaccinations [Online Resource] (2020).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

