Modulation of ERK5 Activity as a Therapeutic Anti-Cancer Strategy
- PMID: 37002872
- PMCID: PMC10108346
- DOI: 10.1021/acs.jmedchem.3c00072
Modulation of ERK5 Activity as a Therapeutic Anti-Cancer Strategy
Abstract
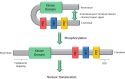
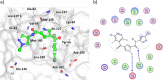
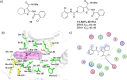
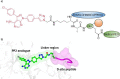
The extracellular signal-regulated kinase 5 (ERK5) signaling pathway is one of four conventional mitogen-activated protein (MAP) kinase pathways. Genetic perturbation of ERK5 has suggested that modulation of ERK5 activity may have therapeutic potential in cancer chemotherapy. This Miniperspective examines the evidence for ERK5 as a drug target in cancer, the structure of ERK5, and the evolution of structurally distinct chemotypes of ERK5 kinase domain inhibitors. The emerging complexities of ERK5 pharmacology are discussed, including the confounding phenomenon of paradoxical ERK5 activation by small-molecule ERK5 inhibitors. The impact of the recent development and biological evaluation of potent and selective bifunctional degraders of ERK5 and future opportunities in ERK modulation are also explored.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information
Miscellaneous

