Context-dependence of T-loop Mediated Long-range RNA Tertiary Interactions
- PMID: 37003469
- PMCID: PMC10152882
- DOI: 10.1016/j.jmb.2023.168070
Context-dependence of T-loop Mediated Long-range RNA Tertiary Interactions
Abstract
The architecture and folding of complex RNAs is governed by a limited set of highly recurrent structural motifs that form long-range tertiary interactions. One of these motifs is the T-loop, which was first identified in tRNA but is broadly distributed across biological RNAs. While the T-loop has been examined in detail in different biological contexts, the various receptors that it interacts with are not as well defined. In this study, we use a cell-based genetic screen in concert with bioinformatic analysis to examine three different, but related, T-loop receptor motifs found in the flavin mononucleotide (FMN) and cobalamin (Cbl) riboswitches. As a host for different T-loop receptors, we employed the env8 class-II Cbl riboswitch, an RNA that uses two T-loop motifs for both folding and supporting the ligand binding pocket. A set of libraries was created in which select nucleotides that participate in the T-loop/T-loop receptor (TL/TLR) interaction were fully randomized. Library members were screened for their ability to support Cbl-dependent expression of a reporter gene. While T-loops appear to be variable in sequence, we find that the functional sequence space is more restricted in the Cbl riboswitch, suggesting that TL/TLR interactions are context dependent. Our data reveal clear sequence signatures for the different types of receptor motifs that align with phylogenic analysis of these motifs in the FMN and Cbl riboswitches. Finally, our data suggest the functional contribution of various nucleobase-mediated long-range interactions within the riboswitch subclass of TL/TLR interactions that are distinct from those found in other RNAs.
Keywords: RNA structure; T-loop; regulatory RNA; structural motif; tertiary interaction.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Interest
R.T.B. serves on the Scientific Advisory Boards of Expansion Therapeutics, SomaLogic and MeiraGTx.
Figures
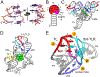
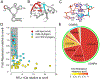
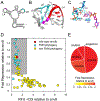
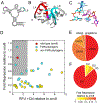

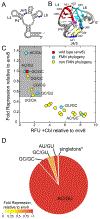
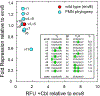
Comment in
-
RNA in the Loop: Probing T-loop/T-loop Receptor Interactions as Mediators of Long-range RNA Contacts that Influence Gene Regulation.J Mol Biol. 2023 May 15;435(10):168087. doi: 10.1016/j.jmb.2023.168087. Epub 2023 Apr 7. J Mol Biol. 2023. PMID: 37030650 No abstract available.
References
-
- Hendrix DK, Brenner SE, Holbrook SR. RNA structural motifs: building blocks of a modular biomolecule. Q Rev Biophys. 2005;38:221–43. - PubMed
-
- Holbrook SR. Structural principles from large RNAs. Annu Rev Biophys. 2008;37:445–64. - PubMed
-
- Butcher SE, Pyle AM. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc Chem Res. 2011;44:1302–11. - PubMed
-
- Batey RT, Rambo RP, Doudna JA. Tertiary Motifs in RNA Structure and Folding. Angew Chem Int Ed Engl. 1999;38:2326–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

