Transcriptomic analysis of albendazole resistance in human diarrheal parasite Giardia duodenalis
- PMID: 37004489
- PMCID: PMC10111952
- DOI: 10.1016/j.ijpddr.2023.03.004
Transcriptomic analysis of albendazole resistance in human diarrheal parasite Giardia duodenalis
Abstract
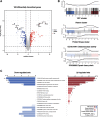
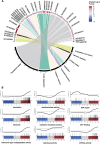
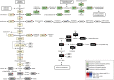
Benzimidazole-2-carbamates (BZ, e.g., albendazole; ALB), which bind β-tubulin to disrupt microtubule polymerization, are one of two primary compound classes used to treat giardiasis. In most parasitic nematodes and fungi, BZ-resistance is caused by β-tubulin mutations and its molecular mode of action (MOA) is well studied. In contrast, in Giardia duodenalis BZ MOA or resistance is less well understood, may involve target-specific and broader impacts including cellular damage and oxidative stress, and its underlying cause is not clearly determined. Previously, we identified acquisition of a single nucleotide polymorphism, E198K, in β-tubulin in ALB-resistant (ALB-R) G. duodenalis WB-1B relative to ALB-sensitive (ALB-S) parental controls. E198K is linked to BZ-resistance in fungi and its allelic frequency correlated with the magnitude of BZ-resistance in G. duodenalis WB-1B. Here, we undertook detailed transcriptomic comparisons of these ALB-S and ALB-R G. duodenalis WB-1B cultures. The primary transcriptional changes with ALB-R in G. duodenalis WB-1B indicated increased protein degradation and turnover, and up-regulation of tubulin, and related genes, associated with the adhesive disc and basal bodies. These findings are consistent with previous observations noting focused disintegration of the disc and associated structures in Giardia duodenalis upon ALB exposure. We also saw transcriptional changes with ALB-R in G. duodenalis WB-1B consistent with prior observations of a shift from glycolysis to arginine metabolism for ATP production and possible changes to aspects of the vesicular trafficking system that require further investigation. Finally, we saw mixed transcriptional changes associated with DNA repair and oxidative stress responses in the G. duodenalis WB-1B line. These changes may be indicative of a role for H2O2 degradation in ALB-R, as has been observed in other G. duodenalis cell cultures. However, they were below the transcriptional fold-change threshold (log2FC > 1) typically employed in transcriptomic analyses and appear to be contradicted in ALB-R G. duodenalis WB-1B by down-regulation of the NAD scavenging and conversion pathways required to support these stress pathways and up-regulation of many highly oxidation sensitive iron-sulphur (FeS) cluster based metabolic enzymes.
Keywords: Albendazole; Drug-resistance; Giardia duodenalis.
Copyright © 2023 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: All authors listed have contributed to the research, have no conflict of interest or competing interests to declare, and have approved the manuscript for submission to IJPDDR. The funders of this study have had no role in its design, data collection and interpretation, and are listed in the manuscript's acknowledgements.
Figures
References
-
- Aguayo-Ortiz R., Méndez-Lucio O., Romo-Mancillas A., Castillo R., Yépez-Mulia L., Medina-Franco J.L., Hernández-Campos A. Molecular basis for benzimidazole resistance from a novel β-tubulin binding site model. J. Mol. Graph. Model. 2013;45:26–37. - PubMed
-
- Alvarado M.E., Wasserman M. Calmodulin expression during Giardia intestinalis differentiation and identification of calmodulin-binding proteins during the trophozoite stage. Parasitol. Res. 2012;110:1371–1380. - PubMed
-
- Andrews S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data. ([Online])
-
- Ankarklev J., Jerlstrom-Hultqvist J., Ringqvist E., Troell K., Svard S.G. Behind the smile: cell biology and disease mechanisms of Giardia species. Nat. Rev. Microbiol. 2010;8:413–422. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

