Dietary Supplementation with Milk Lipids Leads to Suppression of Developmental and Behavioral Phenotypes of Hyperexcitable Drosophila Mutants
- PMID: 37004908
- PMCID: PMC10200772
- DOI: 10.1016/j.neuroscience.2023.03.027
Dietary Supplementation with Milk Lipids Leads to Suppression of Developmental and Behavioral Phenotypes of Hyperexcitable Drosophila Mutants
Abstract
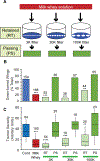
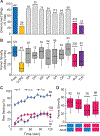
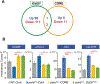
Dietary modifications often have a profound impact on the penetrance and expressivity of neurological phenotypes that are caused by genetic defects. Our previous studies in Drosophila melanogaster revealed that seizure-like phenotypes of gain-of-function voltage-gated sodium (Nav) channel mutants (paraShu, parabss1, and paraGEFS+), as well as other seizure-prone "bang-sensitive" mutants (eas and sda), were drastically suppressed by supplementation of a standard diet with milk whey. In the current study we sought to determine which components of milk whey are responsible for the diet-dependent suppression of their hyperexcitable phenotypes. Our systematic analysis reveals that supplementing the diet with a modest amount of milk lipids (0.26% w/v) mimics the effects of milk whey. We further found that a minor milk lipid component, α-linolenic acid, contributed to the diet-dependent suppression of adult paraShu phenotypes. Given that lipid supplementation during the larval stages effectively suppressed adult paraShu phenotypes, dietary lipids likely modify neural development to compensate for the defects caused by the mutations. Consistent with this notion, lipid feeding fully rescued abnormal dendrite development of class IV sensory neurons in paraShu larvae. Overall, our findings demonstrate that milk lipids are sufficient to ameliorate hyperexcitable phenotypes in Drosophila mutants, providing a foundation for future investigation of the molecular and cellular mechanisms by which dietary lipids modify genetically induced abnormalities in neural development, physiology, and behavior.
Keywords: RNA-sequencing; gene-environment interaction; neuronal development; seizure; voltage-gated sodium channel; α-linolenic acid.
Copyright © 2023 IBRO. Published by Elsevier Ltd. All rights reserved.
Conflict of interest statement
DECLARATIONS OF INTEREST
None
Figures




References
-
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Gruning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D (2018) The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic acids research 46:W537–W544. - PMC - PubMed
-
- Agianian B, Tucker PA, Schouten A, Leonard K, Bullard B, Gros P (2003) Structure of a Drosophila sigma class glutathione S-transferase reveals a novel active site topography suited for lipid peroxidation products. J Mol Biol 326:151–165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

