Multi-tissue DNA methylation aging clocks for sea lions, walruses and seals
- PMID: 37005462
- PMCID: PMC10067968
- DOI: 10.1038/s42003-023-04734-0
Multi-tissue DNA methylation aging clocks for sea lions, walruses and seals
Abstract
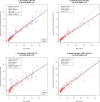
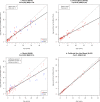
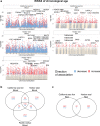
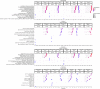
Age determination of wild animals, including pinnipeds, is critical for accurate population assessment and management. For most pinnipeds, current age estimation methodologies utilize tooth or bone sectioning which makes antemortem estimations problematic. We leveraged recent advances in the development of epigenetic age estimators (epigenetic clocks) to develop highly accurate pinniped epigenetic clocks. For clock development, we applied the mammalian methylation array to profile 37,492 cytosine-guanine sites (CpGs) across highly conserved stretches of DNA in blood and skin samples (n = 171) from primarily three pinniped species representing the three phylogenetic families: Otariidae, Phocidae and Odobenidae. We built an elastic net model with Leave-One-Out-Cross Validation (LOOCV) and one with a Leave-One-Species-Out-Cross-Validation (LOSOCV). After identifying the top 30 CpGs, the LOOCV produced a highly correlated (r = 0.95) and accurate (median absolute error = 1.7 years) age estimation clock. The LOSOCV elastic net results indicated that blood and skin clock (r = 0.84) and blood (r = 0.88) pinniped clocks could predict age of animals from pinniped species not used for clock development to within 3.6 and 4.4 years, respectively. These epigenetic clocks provide an improved and relatively non-invasive tool to determine age in skin or blood samples from all pinniped species.
© 2023. The Author(s).
Conflict of interest statement
S.H. is a founder of the non-profit Epigenetic Clock Development Foundation, which plans to license several of his patents from his employer UC Regents. The other authors declare no conflicts of interest.
Figures

References
-
- Garlich-Miller J, Stewart R, Stewart B, Hiltz E. Comparison of mandibular with cemental growth-layer counts for ageing Atlantic walrus (Odobenus rosmarus rosmarus) Can. J. Zool. 1993;71:163–167. doi: 10.1139/z93-022. - DOI
-
- Richard P, Campbell R. Status of the Atlantic walrus, Odobenus rosmarus rosmarus, in Canada. Can. Field Naturalist. 1988;102:337–350.
-
- Previdi M, Smith KL, Polvani LM. Arctic amplification of climate change: a review of underlying mechanisms. Environ. Res. Lett. 2021;16:093003. doi: 10.1088/1748-9326/ac1c29. - DOI
-
- Gulland, F. M. et al. A review of climate change effects on marine mammals in United States waters: past predictions, observed impacts, current research and conservation imperatives. Clim. Chang. Ecol.3, 100054 (2022).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

