Cytoplasmic YAP1-mediated ESCRT-III assembly promotes autophagic cell death and is ubiquitinated by NEDD4L in breast cancer
- PMID: 37005481
- PMCID: PMC10174091
- DOI: 10.1002/cac2.12417
Cytoplasmic YAP1-mediated ESCRT-III assembly promotes autophagic cell death and is ubiquitinated by NEDD4L in breast cancer
Abstract
Background: Nuclear Yes1-associated transcriptional regulator (YAP1) promotes tumor progression. However, the function of cytoplasmic YAP1 in breast cancer cells and its impact on the survival of breast cancer patients remain unclear. Our research aimed to explore the biological function of cytoplasmic YAP1 in breast cancer cells and the possibility of cytoplasmic YAP1 as a predictive marker of breast cancer survival.
Methods: We constructed cell mutant models, including NLS-YAP15SA (nuclear localized), YAP1S94A (incapable of binding to the TEA domain transcription factor family) and YAP1S127D (cytoplasmic localized), and used Cell Counting Kit-8 (CCK-8) assays, 5-ethynyl-2'-deoxyuridine (EdU) incorporation assays, and Western blotting (WB) analysis to detect cell proliferation and apoptosis. The specific mechanism of cytoplasmic YAP1-mediated endosomal sorting complexes required for transport III (ESCRT-III) assembly was studied by co-immunoprecipitation, immunofluorescence staining, and WB analysis. Epigallocatechin gallate (EGCG) was used to simulate YAP1 retention in the cytoplasm in in vitro and in vivo experiments to study the function of cytoplasmic YAP1. YAP1 binding to NEDD4-like E3 ubiquitin protein ligase (NEDD4L) was identified using mass spectrometry and was verified in vitro. Breast tissue microarrays were used to analyze the relationship between cytoplasmic YAP1 expression and the survival of breast cancer patients.
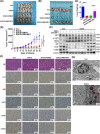
Results: YAP1 was mainly expressed in the cytoplasm in breast cancer cells. Cytoplasmic YAP1 promoted autophagic death of breast cancer cells. Cytoplasmic YAP1 bound to the ESCRT-III complex subunits charged multivesicular body protein 2B (CHMP2B) and vacuolar protein sorting 4 homolog B (VPS4B), promoting assembly of CHMP2B-VPS4B and activating autophagosome formation. EGCG retained YAP1 in the cytoplasm, promoting the assembly of CHMP2B-VPS4B to promote autophagic death of breast cancer cells. YAP1 bound to NEDD4L, and NEDD4L mediated ubiquitination and degradation of YAP1. Breast tissue microarrays revealed that high levels of cytoplasmic YAP1 were beneficial to the survival of breast cancer patients.
Conclusions: Cytoplasmic YAP1 mediated autophagic death of breast cancer cells by promoting assembly of the ESCRT-III complex; furthermore, we established a new breast cancer survival prediction model based on cytoplasmic YAP1 expression.
Keywords: Autophagosome closure; Autophagy; Breast cancer; CHMP2B; Cytoplasmic YAP1; EGCG; ESCRT-III; Hippo pathway; NEDD4L; Ubiquitin; VPS4B.
© 2023 The Authors. Cancer Communications published by John Wiley & Sons Australia, Ltd. on behalf of Sun Yat-sen University Cancer Center.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

