DNA methylome and transcriptome profiling reveal key electrophysiology and immune dysregulation in hypertrophic cardiomyopathy
- PMID: 37005704
- PMCID: PMC10072074
- DOI: 10.1080/15592294.2023.2195307
DNA methylome and transcriptome profiling reveal key electrophysiology and immune dysregulation in hypertrophic cardiomyopathy
Abstract
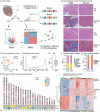
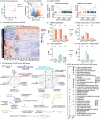
Hypertrophic cardiomyopathy (HCM) is the most common inherited heart disease. However, a detailed DNA methylation (DNAme) landscape has not yet been elucidated. Our study combined DNAme and transcriptome profiles for HCM myocardium and identify aberrant DNAme associated with altered myocardial function in HCM. The transcription of methylation-related genes did not significantly differ between HCM and normal myocardium. Nevertheless, the former had an altered DNAme profile compared with the latter. The hypermethylated and hypomethylated sites in HCM tissues had chromosomal distributions and functional enrichment of correlated genes differing from those of their normal tissue counterparts. The GO analysis of network underlying the genes correlated with DNAme alteration and differentially expressed genes (DEGs) shows functional clusters centred on immune cell function and muscle system processes. In KEGG analysis, only the calcium signalling pathway was enriched either by the genes correlated with changes in DNAme or DEGs. The protein-protein interactions (PPI) underlying the genes altered at both the DNAme and transcriptional highlighted two important functional clusters. One of these was related to the immune response and had the estrogen receptor-encoding ESR1 gene as its node. The other cluster comprised cardiac electrophysiology-related genes. Intelliectin-1 (ITLN1), a component of the innate immune system, was transcriptionally downregulated in HCM and had a hypermethylated site within 1500 bp upstream of the ITLN1 transcription start site. Estimates of immune infiltration demonstrated a relative decline in immune cell population diversity in HCM. A combination of DNAme and transcriptome profiles may help identify and develop new therapeutic targets for HCM.
Keywords: Cardiac electrophysiology; DNA methylation; hypertrophic cardiomyopathy; immune responses; myocardial remodelling; transcriptome.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures

References
-
- Maron BJ, Gardin JM, Flack JM, et al. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–17. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
