T-cell exhaustion signatures characterize the immune landscape and predict HCC prognosis via integrating single-cell RNA-seq and bulk RNA-sequencing
- PMID: 37006257
- PMCID: PMC10050519
- DOI: 10.3389/fimmu.2023.1137025
T-cell exhaustion signatures characterize the immune landscape and predict HCC prognosis via integrating single-cell RNA-seq and bulk RNA-sequencing
Abstract
Background: Hepatocellular carcinoma (HCC), the third most prevalent cause of cancer-related death, is a frequent primary liver cancer with a high rate of morbidity and mortality. T-cell depletion (TEX) is a progressive decline in T-cell function due to continuous stimulation of the TCR in the presence of sustained antigen exposure. Numerous studies have shown that TEX plays an essential role in the antitumor immune process and is significantly associated with patient prognosis. Hence, it is important to gain insight into the potential role of T cell depletion in the tumor microenvironment. The purpose of this study was to develop a trustworthy TEX-based signature using single-cell RNA-seq (scRNA-seq) and high-throughput RNA sequencing, opening up new avenues for evaluating the prognosis and immunotherapeutic response of HCC patients.
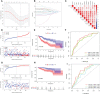
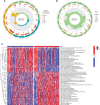
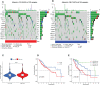
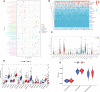
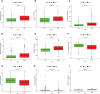
Methods: The International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA) databases were used to download RNA-seq information for HCC patients. The 10x scRNA-seq. data of HCC were downloaded from GSE166635, and UMAP was used for clustering descending, and subgroup identification. TEX-related genes were identified by gene set variance analysis (GSVA) and weighted gene correlation network analysis (WGCNA). Afterward, we established a prognostic TEX signature using LASSO-Cox analysis. External validation was performed in the ICGC cohort. Immunotherapy response was assessed by the IMvigor210, GSE78220, GSE79671, and GSE91061cohorts. In addition, differences in mutational landscape and chemotherapy sensitivity between different risk groups were investigated. Finally, the differential expression of TEX genes was verified by qRT-PCR.
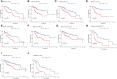
Result: 11 TEX genes were thought to be highly predictive of the prognosis of HCC and substantially related to HCC prognosis. Patients in the low-risk group had a greater overall survival rate than those in the high-risk group, according to multivariate analysis, which also revealed that the model was an independent predictor of HCC. The predictive efficacy of columnar maps created from clinical features and risk scores was strong.
Conclusion: TEX signature and column line plots showed good predictive performance, providing a new perspective for assessing pre-immune efficacy, which will be useful for future precision immuno-oncology studies.
Keywords: HCC; T-cell exhaustion; immunotherapy; machine learning; predictive signature; single-cell RNA-seq; tumor microenvironment.
Copyright © 2023 Chi, Zhao, Yang, Gao, Peng, Zhang, Xie, Song, Xu, Xia, Chen and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

