Polygenic risk score prediction of multiple sclerosis in individuals of South Asian ancestry
- PMID: 37006331
- PMCID: PMC10053643
- DOI: 10.1093/braincomms/fcad041
Polygenic risk score prediction of multiple sclerosis in individuals of South Asian ancestry
Abstract
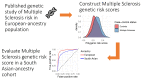
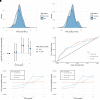
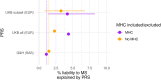
Polygenic risk scores aggregate an individual's burden of risk alleles to estimate the overall genetic risk for a specific trait or disease. Polygenic risk scores derived from genome-wide association studies of European populations perform poorly for other ancestral groups. Given the potential for future clinical utility, underperformance of polygenic risk scores in South Asian populations has the potential to reinforce health inequalities. To determine whether European-derived polygenic risk scores underperform at multiple sclerosis prediction in a South Asian-ancestry population compared with a European-ancestry cohort, we used data from two longitudinal genetic cohort studies: Genes & Health (2015-present), a study of ∼50 000 British-Bangladeshi and British-Pakistani individuals, and UK Biobank (2006-present), which is comprised of ∼500 000 predominantly White British individuals. We compared individuals with and without multiple sclerosis in both studies (Genes & Health: N Cases = 42, N Control = 40 490; UK Biobank: N Cases = 2091, N Control = 374 866). Polygenic risk scores were calculated using clumping and thresholding with risk allele effect sizes obtained from the largest multiple sclerosis genome-wide association study to date. Scores were calculated with and without the major histocompatibility complex region, the most influential locus in determining multiple sclerosis risk. Polygenic risk score prediction was evaluated using Nagelkerke's pseudo-R 2 metric adjusted for case ascertainment, age, sex and the first four genetic principal components. We found that, as expected, European-derived polygenic risk scores perform poorly in the Genes & Health cohort, explaining 1.1% (including the major histocompatibility complex) and 1.5% (excluding the major histocompatibility complex) of disease risk. In contrast, multiple sclerosis polygenic risk scores explained 4.8% (including the major histocompatibility complex) and 2.8% (excluding the major histocompatibility complex) of disease risk in European-ancestry UK Biobank participants. These findings suggest that polygenic risk score prediction of multiple sclerosis based on European genome-wide association study results is less accurate in a South Asian population. Genetic studies of ancestrally diverse populations are required to ensure that polygenic risk scores can be useful across ancestries.
Keywords: ethnicity; genetics; multiple sclerosis.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
The authors report no competing interests.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
