High-fat intake reshapes the circadian transcriptome profile and metabolism in murine meibomian glands
- PMID: 37006922
- PMCID: PMC10062204
- DOI: 10.3389/fnut.2023.1146916
High-fat intake reshapes the circadian transcriptome profile and metabolism in murine meibomian glands
Abstract
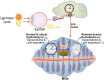
Background: Nutritional and food components reshape the peripheral clock and metabolism. However, whether food challenges affect the circadian clock and metabolism of meibomian glands (MGs) has not been fully explored. This study was designed to analyze alterations in the rhythmic transcriptome and metabolism of MGs of murine fed a balanced diet or a high-fat diet (HFD).
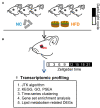
Methods: Male C57BL/6J mice were maintained on a 12/12 h light/dark cycle and fed ad libitum on normal chow (NC) or HFD for 4 weeks. MGs were collected from sacrificed animals at 3-h intervals throughout a 24-h circadian cycle. The circadian transcriptome of MGs was analyzed via bioinformatics approaches using high-throughput RNA sequencing (RNA-seq). In addition, circadian oscillations of lipid components in MGs were analyzed.
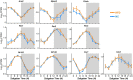
Results: Meibomian glands displayed robust transcriptome rhythmicity. HFD feeding significantly altered the circadian transcriptome profile of MGs-including composition and phase-and spatiotemporally affected the enriched signaling pathways. In addition, HFD feeding significantly altered the normal rhythmic oscillations of lipid components in MGs.
Conclusion: Our data show that HFD significantly affects MGs' rhythmicity, which reveals a high sensitivity of MGs' clocks to lipid composition in food.
Keywords: RNA-seq; bioinformatics; circadian rhythm; high-fat diet; meibomian gland; metabolic dysfunction; transcriptome.
Copyright © 2023 Zou, Liu, Si, Huang, Qi, Pei, Lu, Huang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources

