A comprehensive pan-cancer analysis unveiling the oncogenic effect of plant homeodomain finger protein 14 (PHF14) in human tumors
- PMID: 37007943
- PMCID: PMC10061232
- DOI: 10.3389/fgene.2023.1073138
A comprehensive pan-cancer analysis unveiling the oncogenic effect of plant homeodomain finger protein 14 (PHF14) in human tumors
Abstract
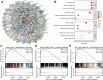
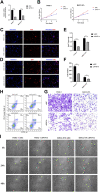
The plant homeodomain (PHD) finger refers to a protein motif that plays a key role in the recognition and translation of histone modification marks by promoting gene transcriptional activation and silencing. As an important member of the PHD family, the plant homeodomain finger protein 14 (PHF14) affects the biological behavior of cells as a regulatory factor. Several emerging studies have demonstrated that PHF14 expression is closely associated with the development of some cancers, but there is still no feasible pan-cancer analysis. Based on existing datasets from the Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO), we performed a systematic analysis of the oncogenic role of the PHF14 gene in 33 human cancers. The expression level of PHF14 was significantly different between different types of tumors and adjacent normal tissues, and the expression or genetic alteration of PHF14 gene was closely related to the prognosis of most cancer patients. Levels of cancer-associated fibroblasts (CAFs) infiltration in various cancer types were also observed to correlate with PHF14 expression. In some tumors, PFH14 may play a role in tumor immunity by regulating the expression levels of immune checkpoint genes. In addition, the results of enrichment analysis showed that the main biological activities of PHF14 were related to various signaling pathways or chromatin complex effects. In conclusion, our pan-cancer research shows that the expression level of PHF14 is closely related to the carcinogenesis and prognosis of certain tumors, which needs to be further verified by more experiments and more in-depth mechanism exploration.
Keywords: PHF14; cancer; carcinogenesis; gene expression; pan-cancer analysis; prognosis.
Copyright © 2023 Cao, Zhan, Wu, Kuang, Mo, Liu and Dai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Deciphering the dual roles of PHD finger proteins from oncogenic drivers to tumor suppressors.Front Cell Dev Biol. 2024 May 15;12:1403396. doi: 10.3389/fcell.2024.1403396. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38813086 Free PMC article. Review.
-
Depletion of PHF14, a novel histone-binding protein gene, causes neonatal lethality in mice due to respiratory failure.Acta Biochim Biophys Sin (Shanghai). 2013 Aug;45(8):622-33. doi: 10.1093/abbs/gmt055. Epub 2013 May 20. Acta Biochim Biophys Sin (Shanghai). 2013. PMID: 23688586
-
A novel PHD-finger protein 14/KIF4A complex overexpressed in lung cancer is involved in cell mitosis regulation and tumorigenesis.Oncotarget. 2017 Mar 21;8(12):19684-19698. doi: 10.18632/oncotarget.14962. Oncotarget. 2017. PMID: 28160558 Free PMC article.
-
Pulsed SILAC-based proteomic analysis unveils hypoxia- and serum starvation-induced de novo protein synthesis with PHD finger protein 14 (PHF14) as a hypoxia sensitive epigenetic regulator in cell cycle progression.Oncotarget. 2019 Mar 15;10(22):2136-2150. doi: 10.18632/oncotarget.26669. eCollection 2019 Mar 15. Oncotarget. 2019. PMID: 31040906 Free PMC article.
-
A pan-cancer analysis targeting the oncogenic role of ATP-binding cassette transporter A1 in human tumors.Front Oncol. 2025 Apr 14;15:1513992. doi: 10.3389/fonc.2025.1513992. eCollection 2025. Front Oncol. 2025. PMID: 40297807 Free PMC article. Review.
Cited by
-
Deciphering the dual roles of PHD finger proteins from oncogenic drivers to tumor suppressors.Front Cell Dev Biol. 2024 May 15;12:1403396. doi: 10.3389/fcell.2024.1403396. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38813086 Free PMC article. Review.
-
A sequencing-based screening method identifies regulators of EGFR signaling from nonviable mutants in Caenorhabditis elegans.Sci Signal. 2025 Feb 25;18(875):eadp9377. doi: 10.1126/scisignal.adp9377. Epub 2025 Feb 25. Sci Signal. 2025. PMID: 39999212 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

