Identification of the cuproptosis-related hub genes and therapeutic agents for sarcopenia
- PMID: 37007946
- PMCID: PMC10063920
- DOI: 10.3389/fgene.2023.1136763
Identification of the cuproptosis-related hub genes and therapeutic agents for sarcopenia
Abstract
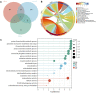
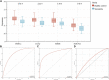
Background: Along with acceleration of population aging, the increasing prevalence of sarcopenia has posed a heavy burden on families as well as society. In this context, it is of great significance to diagnose and intervene sarcopenia as early as possible. Recent evidence has indicated the role of cuproptosis in the development of sarcopenia. In this study, we aimed to seek the key cuproptosis-related genes that can be used for identification and intervention of sarcopenia. Methods: The GSE111016 dataset was retrieved from GEO. The 31 cuproptosis-related genes (CRGs) were obtained from previous published studies. The differentially expressed genes (DEGs) and Weighed gene co-expression network analysis (WGCNA) were subsequently analyzed. The core hub genes were acquired by the intersection of DEGs, WGCNA and CRGs. Through logistic regression analysis, we established a diagnostic model of sarcopenia based on the selected biomarkers and was validated in muscle samples from GSE111006 and GSE167186. In addition, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analysis were performed on these genes. Furthermore, the gene set enrichment analysis (GSEA), and immune cell infiltration were also conducted on the identified core genes. Finally, we screened the potential drugs targeting the potential biomarkers of sarcopenia. Results: A total of 902 DEGs and WGCNA containing 1,281 significant genes were preliminarily selected. Intersection of DEGs, WGCNA and CRGs yielded four core genes (PDHA1, DLAT, PDHB, and NDUFC1) as potential biomarkers for the prediction of sarcopenia. The predictive model was established and validated with high AUC values. KEGG pathway and Gene Ontology biological analysis indicated these core genes may play a crucial role in energy metabolism in mitochondria, oxidation process, and aging-related degenerative diseases. In addition, the immune cells may be involved in the development of sarcopenia through mitochondrial metabolism. Finally, metformin was identified as a promising strategy of sarcopenia treatment via targeting NDUFC1. Conclusion: The four cuproptosis-related genes PDHA1, DLAT, PDHB and NDUFC1 may be the diagnostic biomarkers for sarcopenia, and metformin holds great potential to be developed as a therapy for sarcopenia. These outcomes provide new insights for better understanding of sarcopenia and innovative therapeutic approaches.
Keywords: bioinformatics; cuproptosis; metformin; mitochondrial metabolism; sarcopenia.
Copyright © 2023 Zhu, Chen, Geng, Li, Li, Yuan and Jiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Alvarez-Calderon F., Gregory M. A., Pham-Danis C., DeRyckere D., Stevens B. M., Zaberezhnyy V., et al. (2015). Tyrosine kinase inhibition in leukemia induces an altered metabolic state sensitive to mitochondrial perturbations. Clin. Cancer Res. 21 (6), 1360–1372. 10.1158/1078-0432.Ccr-14-2146 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

