EpiCRISPR targeted methylation of Arx gene initiates transient switch of mouse pancreatic alpha to insulin-producing cells
- PMID: 37008919
- PMCID: PMC10063207
- DOI: 10.3389/fendo.2023.1134478
EpiCRISPR targeted methylation of Arx gene initiates transient switch of mouse pancreatic alpha to insulin-producing cells
Abstract
Introduction: Beta cell dysfunction by loss of beta cell identity, dedifferentiation, and the presence of polyhormonal cells are main characteristics of diabetes. The straightforward strategy for curing diabetes implies reestablishment of pancreatic beta cell function by beta cell replacement therapy. Aristaless-related homeobox (Arx) gene encodes protein which plays an important role in the development of pancreatic alpha cells and is a main target for changing alpha cell identity.
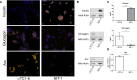
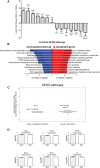
Results: In this study we used CRISPR/dCas9-based epigenetic tools for targeted hypermethylation of Arx gene promoter and its subsequent suppression in mouse pancreatic αTC1-6 cell line. Bisulfite sequencing and methylation profiling revealed that the dCas9-Dnmt3a3L-KRAB single chain fusion constructs (EpiCRISPR) was the most efficient. Epigenetic silencing of Arx expression was accompanied by an increase in transcription of the insulin gene (Ins2) mRNA on 5th and 7th post-transfection day, quantified by both RT-qPCR and RNA-seq. Insulin production and secretion was determined by immunocytochemistry and ELISA assay, respectively. Eventually, we were able to induce switch of approximately 1% of transiently transfected cells which were able to produce 35% more insulin than Mock transfected alpha cells.
Conclusion: In conclusion, we successfully triggered a direct, transient switch of pancreatic alpha to insulin-producing cells opening a future research on promising therapeutic avenue for diabetes management.
Keywords: Arx gene; CRISPR/dCas9; diabetes; epigenetic editing; pancreatic alpha cells; targeted DNA methylation.
Copyright © 2023 Đorđević, Stepper, Feuerstein-Akgoz, Gerhauser, Paunović, Tolić, Rajić, Dinić, Uskoković, Grdović, Mihailović, Jurkowska, Jurkowski, Jovanović and Vidaković.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

