Integrative multi-omics approaches to explore immune cell functions: Challenges and opportunities
- PMID: 37009227
- PMCID: PMC10060681
- DOI: 10.1016/j.isci.2023.106359
Integrative multi-omics approaches to explore immune cell functions: Challenges and opportunities
Abstract
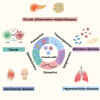
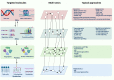
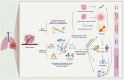
As modern biological sciences evolve from investigation of individual molecules and pathways to growing emphasis on global and systems-based processes, increasing efforts have focused on combining the study of genomics with that of the other omics technologies, including epigenomics, transcriptomics, quantitative proteomics, global analyses of post-translational modifications (PTMs) and metabolomics, to characterize specific biological or pathological processes. In addition, emerging genome-wide functional screening technologies further help researchers identify key regulators of immune functions. Derived from these multi-omics technologies, single cell sequencing analysis on multiple layers offers an overview of intra-tissue or intra-organ immune cell heterogeneity. In this review, we summarize advances in multi-omics tools to explore immune cell functions and applications of these multi-omics approaches in the analysis of clinical immune disorders, aiming to provide an outlook on the potential opportunities and challenges that these technologies pose in future investigation in the field of immunology.
Keywords: Bioinformatics; Cell biology; Immunology.
© 2023 The Authors.
Conflict of interest statement
No conflict of interest is declared.
Figures


Similar articles
-
Omics-Based Investigations of Breast Cancer.Molecules. 2023 Jun 14;28(12):4768. doi: 10.3390/molecules28124768. Molecules. 2023. PMID: 37375323 Free PMC article. Review.
-
[Multi-omics technology and its applications to life sciences: a review].Sheng Wu Gong Cheng Xue Bao. 2022 Oct 25;38(10):3581-3593. doi: 10.13345/j.cjb.220724. Sheng Wu Gong Cheng Xue Bao. 2022. PMID: 36305394 Review. Chinese.
-
Integrative Analysis of Multi-omics Data for Discovery and Functional Studies of Complex Human Diseases.Adv Genet. 2016;93:147-90. doi: 10.1016/bs.adgen.2015.11.004. Epub 2016 Jan 25. Adv Genet. 2016. PMID: 26915271 Free PMC article. Review.
-
"Omics" in pharmaceutical research: overview, applications, challenges, and future perspectives.Chin J Nat Med. 2015 Jan;13(1):3-21. doi: 10.1016/S1875-5364(15)60002-4. Chin J Nat Med. 2015. PMID: 25660284 Review.
-
Multi-omics study for interpretation of genome-wide association study.J Hum Genet. 2021 Jan;66(1):3-10. doi: 10.1038/s10038-020-00842-5. Epub 2020 Sep 18. J Hum Genet. 2021. PMID: 32948838 Review.
Cited by
-
Application of machine learning for mass spectrometry-based multi-omics in thyroid diseases.Front Mol Biosci. 2024 Dec 17;11:1483326. doi: 10.3389/fmolb.2024.1483326. eCollection 2024. Front Mol Biosci. 2024. PMID: 39741929 Free PMC article. Review.
-
Chronic Pelvic Pain, Vulvar Pain Disorders, and Proteomics Profiles: New Discoveries, New Hopes.Biomedicines. 2023 Dec 19;12(1):1. doi: 10.3390/biomedicines12010001. Biomedicines. 2023. PMID: 38275362 Free PMC article. Review.
-
Bone Trans-omics: Integrating Omics to Unveil Mechanistic Molecular Networks Regulating Bone Biology and Disease.Curr Osteoporos Rep. 2023 Oct;21(5):493-502. doi: 10.1007/s11914-023-00812-8. Epub 2023 Jul 6. Curr Osteoporos Rep. 2023. PMID: 37410317 Free PMC article. Review.
-
Predicting Alzheimer's disease subtypes and understanding their molecular characteristics in living patients with transcriptomic trajectory profiling.Alzheimers Dement. 2025 Jan;21(1):e14241. doi: 10.1002/alz.14241. Epub 2025 Jan 15. Alzheimers Dement. 2025. PMID: 39812331 Free PMC article.
-
T cells in the microenvironment of solid pediatric tumors: the case of neuroblastoma.Front Immunol. 2025 Feb 28;16:1544137. doi: 10.3389/fimmu.2025.1544137. eCollection 2025. Front Immunol. 2025. PMID: 40092980 Free PMC article. Review.
References
-
- Johnson Chavarria E.M. A primer of human genetics. Yale J. Biol. Med. 2016;89:603.
-
- Tan H., Yang K., Li Y., Shaw T.I., Wang Y., Blanco D.B., Wang X., Cho J.H., Wang H., Rankin S., et al. Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity. 2017;46:488–503. doi: 10.1016/j.immuni.2017.02.010. - DOI - PMC - PubMed
-
- Bakker O.B., Aguirre-Gamboa R., Sanna S., Oosting M., Smeekens S.P., Jaeger M., Zorro M., Võsa U., Withoff S., Netea-Maier R.T., et al. Integration of multi-omics data and deep phenotyping enables prediction of cytokine responses. Nat. Immunol. 2018;19:776–786. doi: 10.1038/s41590-018-0121-3. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

