Identification of a pediatric acute hypoxemic respiratory failure signature in peripheral blood leukocytes at 24 hours post-ICU admission with machine learning
- PMID: 37009294
- PMCID: PMC10063855
- DOI: 10.3389/fped.2023.1159473
Identification of a pediatric acute hypoxemic respiratory failure signature in peripheral blood leukocytes at 24 hours post-ICU admission with machine learning
Abstract
Background: There is no generalizable transcriptomics signature of pediatric acute respiratory distress syndrome. Our goal was to identify a whole blood differential gene expression signature for pediatric acute hypoxemic respiratory failure (AHRF) using transcriptomic microarrays within twenty-four hours of diagnosis. We used publicly available human whole-blood gene expression arrays of a Berlin-defined pediatric acute respiratory distress syndrome (GSE147902) cohort and a sepsis-triggered AHRF (GSE66099) cohort within twenty-four hours of diagnosis and compared those children with a PaO2/FiO2 < 200 to those with a PaO2/FiO2 ≥ 200.
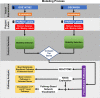
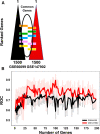
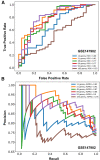
Results: We used stability selection, a bootstrapping method of 100 simulations using logistic regression as a classifier, to select differentially expressed genes associated with a PaO2/FiO2 < 200 vs. PaO2/FiO2 ≥ 200. The top-ranked genes that contributed to the AHRF signature were selected in each dataset. Genes common to both of the top 1,500 ranked gene lists were selected for pathway analysis. Pathway and network analysis was performed using the Pathway Network Analysis Visualizer (PANEV) and Reactome was used to perform an over-representation gene network analysis of the top-ranked genes common to both cohorts. Changes in metabolic pathways involved in energy balance, fundamental cellular processes such as protein translation, mitochondrial function, oxidative stress, immune signaling, and inflammation are differentially regulated early in pediatric ARDS and sepsis-induced AHRF compared to both healthy controls and to milder acute hypoxemia. Specifically, fundamental pathways related to the severity of hypoxemia emerged and included (1) ribosomal and eukaryotic initiation of factor 2 (eIF2) regulation of protein translation and (2) the nutrient, oxygen, and energy sensing pathway, mTOR, activated via PI3K/AKT signaling.
Conclusions: Cellular energetics and metabolic pathways are important mechanisms to consider to further our understanding of the heterogeneity and underlying pathobiology of moderate and severe pediatric acute respiratory distress syndrome. Our findings are hypothesis generating and support the study of metabolic pathways and cellular energetics to understand heterogeneity and underlying pathobiology of moderate and severe acute hypoxemic respiratory failure in children.
Keywords: acute respiratory distress syndrome; gene expression profiling; machine learning; mechanical ventilation; pediatric; transcriptomics.
© 2023 Grunwell, Rad, Ripple, Yehya, Wong and Kamaleswaran.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Sinha P, Delucchi KL, Thompson BT, McAuley DF, Matthay MA, Calfee CS, et al. Latent class analysis of ARDS subphenotypes: a secondary analysis of the statins for acutely injured lungs from sepsis (SAILS) study. Intensive Care Med. (2018) 44(11):1859–69. 10.1007/s00134-018-5378-3 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

