Proteomic characterization of extracellular vesicles released by third stage larvae of the zoonotic parasite Anisakis pegreffii (Nematoda: Anisakidae)
- PMID: 37009516
- PMCID: PMC10050594
- DOI: 10.3389/fcimb.2023.1079991
Proteomic characterization of extracellular vesicles released by third stage larvae of the zoonotic parasite Anisakis pegreffii (Nematoda: Anisakidae)
Abstract
Introduction: Anisakis pegreffii is a sibling species within the A. simplex (s.l.) complex requiring marine homeothermic (mainly cetaceans) and heterothermic (crustaceans, fish, and cephalopods) organisms to complete its life cycle. It is also a zoonotic species, able to accidentally infect humans (anisakiasis). To investigate the molecular signals involved in this host-parasite interaction and pathogenesis, the proteomic composition of the extracellular vesicles (EVs) released by the third-stage larvae (L3) of A. pegreffii, was characterized.
Methods: Genetically identified L3 of A. pegreffii were maintained for 24 h at 37°C and EVs were isolated by serial centrifugation and ultracentrifugation of culture media. Proteomic analysis was performed by Shotgun Analysis.
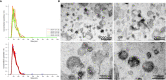
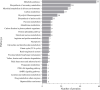
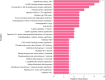
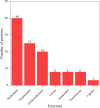
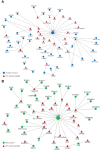
Results and discussion: EVs showed spherical shaped structure (size 65-295 nm). Proteomic results were blasted against the A. pegreffii specific transcriptomic database, and 153 unique proteins were identified. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analysis predicted several proteins belonging to distinct metabolic pathways. The similarity search employing selected parasitic nematodes database revealed that proteins associated with A. pegreffii EVs might be involved in parasite survival and adaptation, as well as in pathogenic processes. Further, a possible link between the A. pegreffii EVs proteins versus those of human and cetaceans' hosts, were predicted by using HPIDB database. The results, herein described, expand knowledge concerning the proteins possibly implied in the host-parasite interactions between this parasite and its natural and accidental hosts.
Keywords: Anisakis pegreffii; allergenic proteins; extracellular vesicles; heat shock proteins; metalloproteases; proteomics; third stage larvae; zoonotic parasite.
Copyright © 2023 Palomba, Rughetti, Mignogna, Castrignanò, Rahimi, Masuelli, Napoletano, Pinna, Giorgi, Santoro, Schininà, Maras and Mattiucci.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

