A frameshift in Yersinia pestis rcsD alters canonical Rcs signalling to preserve flea-mammal plague transmission cycles
- PMID: 37010269
- PMCID: PMC10191623
- DOI: 10.7554/eLife.83946
A frameshift in Yersinia pestis rcsD alters canonical Rcs signalling to preserve flea-mammal plague transmission cycles
Abstract
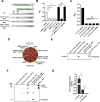
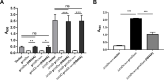
Multiple genetic changes in the enteric pathogen Yersinia pseudotuberculosis have driven the emergence of Yesinia pestis, the arthropod-borne, etiological agent of plague. These include developing the capacity for biofilm-dependent blockage of the flea foregut to enable transmission by flea bite. Previously, we showed that pseudogenization of rcsA, encoding a component of the Rcs signalling pathway, is an important evolutionary step facilitating Y. pestis flea-borne transmission. Additionally, rcsD, another important gene in the Rcs system, harbours a frameshift mutation. Here, we demonstrated that this rcsD mutation resulted in production of a small protein composing the C-terminal RcsD histidine-phosphotransferase domain (designated RcsD-Hpt) and full-length RcsD. Genetic analysis revealed that the rcsD frameshift mutation followed the emergence of rcsA pseudogenization. It further altered the canonical Rcs phosphorylation signal cascade, fine-tuning biofilm production to be conducive with retention of the pgm locus in modern lineages of Y. pestis. Taken together, our findings suggest that a frameshift mutation in rcsD is an important evolutionary step that fine-tuned biofilm production to ensure perpetuation of flea-mammal plague transmission cycles.
Keywords: Rcs phosphorelay system; Yersinia pestis; biofilm; flea; frameshift; infectious disease; microbiology; rcsD.
Plain language summary
Yersinia pestis, the agent responsible for the plague, emerged 6,000 to 7,000 years ago from Yersinia pseudotuberculosis, another type of bacteria which still exists today. Although they are highly similar genetically, these two species are strikingly different. While Y. pseudotuberculosis spreads via food and water and causes mild stomach distress, Y. pestis uses fleas to infect new hosts and has killed millions. A small set of genetic changes has contributed to the emergence of Y. pestis by allowing it to thrive inside a flea and maximise its transmission. In particular, some of these mutations have led to the bacteria being able to come together to form a sticky layer that adheres to the gut of the insect, with this ‘biofilm’ stopping the flea from feeding on blood. The starving flea keeps trying to feed, and with each bite comes another opportunity for Y. pestis to jump host. However, it remains unclear exactly how the mutations have influenced biofilm formation to allow for this new transmission mechanism to take place. To examine this phenomenon, Guo et al. focused on rcsD, a gene that codes for a component of the signalling system that controls biofilm creation. In Y. pestis this sequence has been mutated to become a ‘pseudogene’, a type of sequence which is often thought to be non-functional. However, the experiments showed that, in Y. pestis, rcsD could produce small amounts of a full-length RcsD protein similar to the one found in Y. pseudotuberculosis. However, the gene mostly produces a short ‘RcsD-Hpt’ protein that can, in turn, alter the expression of many genes, including those that decrease biofilm formation. This may prove to be beneficial for Y. pestis, for example when the bacteria switches from living in fleas to living in humans, where it does not require a biofilm. Guo et al. further investigated the impact of rcsD becoming a pseudogene inY. pestis, showing that if normal amounts of the full-length RcsD protein are produced, the bacteria quickly lose the gene that allows them to form biofilm in fleas, and cause disease in humans. In fact, additional analyses revealed that all sequenced strains of ancient and modern Y. pestis bacteria can produce RcsD-Hpt, even if they do not carry the same exact rcsD mutation. Overall, these results indicate that rcsD turning into a pseudogene marked an important step in the emergence of Y. pestis strains that can cause lasting plague outbreaks. They also point towards pseudogenes having more important roles in evolution than previously thought.
© 2023, Guo, Yan, Yang et al.
Conflict of interest statement
XG, HY, WY, ZY, VV, DZ, YS No competing interests declared
Figures









Update of
- doi: 10.1101/2022.10.24.512971
Similar articles
-
Role of the Yersinia pestis phospholipase D (Ymt) in the initial aggregation step of biofilm formation in the flea.mBio. 2024 Jun 12;15(6):e0012424. doi: 10.1128/mbio.00124-24. Epub 2024 May 9. mBio. 2024. PMID: 38722159 Free PMC article.
-
Serotype differences and lack of biofilm formation characterize Yersinia pseudotuberculosis infection of the Xenopsylla cheopis flea vector of Yersinia pestis.J Bacteriol. 2006 Feb;188(3):1113-9. doi: 10.1128/JB.188.3.1113-1119.2006. J Bacteriol. 2006. PMID: 16428415 Free PMC article.
-
Biovar-related differences apparent in the flea foregut colonization phenotype of distinct Yersinia pestis strains do not impact transmission efficiency.Parasit Vectors. 2020 Jul 1;13(1):335. doi: 10.1186/s13071-020-04207-x. Parasit Vectors. 2020. PMID: 32611387 Free PMC article.
-
Yersinia pestis biofilm in the flea vector and its role in the transmission of plague.Curr Top Microbiol Immunol. 2008;322:229-48. doi: 10.1007/978-3-540-75418-3_11. Curr Top Microbiol Immunol. 2008. PMID: 18453279 Free PMC article. Review.
-
Molecular and Genetic Mechanisms That Mediate Transmission of Yersinia pestis by Fleas.Biomolecules. 2021 Feb 3;11(2):210. doi: 10.3390/biom11020210. Biomolecules. 2021. PMID: 33546271 Free PMC article. Review.
Cited by
-
The CpxAR signaling system confers a fitness advantage for flea gut colonization by the plague bacillus.J Bacteriol. 2024 Sep 19;206(9):e0017324. doi: 10.1128/jb.00173-24. Epub 2024 Aug 19. J Bacteriol. 2024. PMID: 39158280 Free PMC article.
-
Characterization of an aspartate aminotransferase encoded by YPO0623 with frequent nonsense mutations in Yersinia pestis.Front Cell Infect Microbiol. 2023 Nov 28;13:1288371. doi: 10.3389/fcimb.2023.1288371. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38089818 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

