FLASH-TB: an Application of Next-Generation CRISPR to Detect Drug Resistant Tuberculosis from Direct Sputum
- PMID: 37010411
- PMCID: PMC10117099
- DOI: 10.1128/jcm.01634-22
FLASH-TB: an Application of Next-Generation CRISPR to Detect Drug Resistant Tuberculosis from Direct Sputum
Abstract
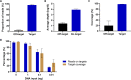
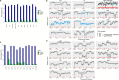
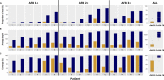
Offering patients with tuberculosis (TB) an optimal and timely treatment regimen depends on the rapid detection of Mycobacterium tuberculosis (Mtb) drug resistance from clinical samples. Finding Low Abundance Sequences by Hybridization (FLASH) is a technique that harnesses the efficiency, specificity, and flexibility of the Cas9 enzyme to enrich targeted sequences. Here, we used FLASH to amplify 52 candidate genes probably associated with resistance to first- and second-line drugs in the Mtb reference strain (H37Rv), then detect drug resistance mutations in cultured Mtb isolates, and in sputum samples. 92% of H37Rv reads mapped to Mtb targets, with 97.8% of target regions covered at a depth ≥ 10X. Among cultured isolates, FLASH-TB detected the same 17 drug resistance mutations as whole genome sequencing (WGS) did, but with much greater depth. Among the 16 sputum samples, FLASH-TB increased recovery of Mtb DNA compared with WGS (from 1.4% [IQR 0.5-7.5] to 33% [IQR 4.6-66.3]) and average depth reads of targets (from 6.3 [IQR 3.8-10.5] to 1991 [IQR 254.4-3623.7]). FLASH-TB identified Mtb complex in all 16 samples based on IS1081 and IS6110 copies. Drug resistance predictions for 15/16 (93.7%) clinical samples were highly concordant with phenotypic DST for isoniazid, rifampicin, amikacin, and kanamycin [15/15 (100%)], ethambutol [12/15 (80%)] and moxifloxacin [14/15 (93.3%)]. These results highlighted the potential of FLASH-TB for detecting Mtb drug resistance from sputum samples.
Keywords: CRISPR; FLASH; Mycobacterium tuberculosis; RNA guide; drug resistance; sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Günther G, Lange C, Alexandru S, Altet N, Avsar K, Bang D, Barbuta R, Bothamley G, Ciobanu A, Crudu V, Danilovits M, Dedicoat M, Duarte R, Gualano G, Kunst H, de Lange W, Leimane V, Magis-Escurra C, McLaughlin A-M, Muylle I, Polcová V, Popa C, Rumetshofer R, Skrahina A, Solodovnikova V, Spinu V, Tiberi S, Viiklepp P, van Leth F, for TBNET . 2016. Treatment outcomes in multidrug-resistant tuberculosis. N Engl J Med 375:1103–1105. doi:10.1056/NEJMc1603274. - DOI - PubMed
-
- World Health Organization. 2022. Global tuberculosis report 2022. WHO.
-
- World Health Organization. 2022. Rapid communication: key changes to the treatment of drug-resistant tuberculosis. WHO.
-
- World Health Organization. 2021. Global tuberculosis report 2021. WHO.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

