Sterol methyltransferases in uncultured bacteria complicate eukaryotic biomarker interpretations
- PMID: 37012227
- PMCID: PMC10070321
- DOI: 10.1038/s41467-023-37552-3
Sterol methyltransferases in uncultured bacteria complicate eukaryotic biomarker interpretations
Abstract
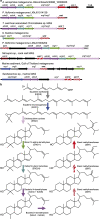
Sterane molecular fossils are broadly interpreted as eukaryotic biomarkers, although diverse bacteria also produce sterols. Steranes with side-chain methylations can act as more specific biomarkers if their sterol precursors are limited to particular eukaryotes and are absent in bacteria. One such sterane, 24-isopropylcholestane, has been attributed to demosponges and potentially represents the earliest evidence for animals on Earth, but enzymes that methylate sterols to give the 24-isopropyl side-chain remain undiscovered. Here, we show that sterol methyltransferases from both sponges and yet-uncultured bacteria function in vitro and identify three methyltransferases from symbiotic bacteria each capable of sequential methylations resulting in the 24-isopropyl sterol side-chain. We demonstrate that bacteria have the genomic capacity to synthesize side-chain alkylated sterols, and that bacterial symbionts may contribute to 24-isopropyl sterol biosynthesis in demosponges. Together, our results suggest bacteria should not be dismissed as potential contributing sources of side-chain alkylated sterane biomarkers in the rock record.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

