Transcriptomic data meta-analysis reveals common and injury model specific gene expression changes in the regenerating zebrafish heart
- PMID: 37012284
- PMCID: PMC10070245
- DOI: 10.1038/s41598-023-32272-6
Transcriptomic data meta-analysis reveals common and injury model specific gene expression changes in the regenerating zebrafish heart
Abstract
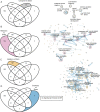
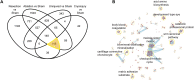
Zebrafish have the capacity to fully regenerate the heart after an injury, which lies in sharp contrast to the irreversible loss of cardiomyocytes after a myocardial infarction in humans. Transcriptomics analysis has contributed to dissect underlying signaling pathways and gene regulatory networks in the zebrafish heart regeneration process. This process has been studied in response to different types of injuries namely: ventricular resection, ventricular cryoinjury, and genetic ablation of cardiomyocytes. However, there exists no database to compare injury specific and core cardiac regeneration responses. Here, we present a meta-analysis of transcriptomic data of regenerating zebrafish hearts in response to these three injury models at 7 days post injury (7dpi). We reanalyzed 36 samples and analyzed the differentially expressed genes (DEG) followed by downstream Gene Ontology Biological Processes (GO:BP) analysis. We found that the three injury models share a common core of DEG encompassing genes involved in cell proliferation, the Wnt signaling pathway and genes that are enriched in fibroblasts. We also found injury-specific gene signatures for resection and genetic ablation, and to a lower extent the cryoinjury model. Finally, we present our data in a user-friendly web interface that displays gene expression signatures across different injury types and highlights the importance to consider injury-specific gene regulatory networks when interpreting the results related to cardiac regeneration in the zebrafish. The analysis is freely available at: https://mybinder.org/v2/gh/MercaderLabAnatomy/PUB_Botos_et_al_2022_shinyapp_binder/HEAD?urlpath=shiny/bus-dashboard/ .
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures