Genetic effects on the timing of parturition and links to fetal birth weight
- PMID: 37012456
- PMCID: PMC10101852
- DOI: 10.1038/s41588-023-01343-9
Genetic effects on the timing of parturition and links to fetal birth weight
Erratum in
-
Author Correction: Genetic effects on the timing of parturition and links to fetal birth weight.Nat Genet. 2023 Jul;55(7):1250. doi: 10.1038/s41588-023-01412-z. Nat Genet. 2023. PMID: 37165137 Free PMC article. No abstract available.
Abstract
The timing of parturition is crucial for neonatal survival and infant health. Yet, its genetic basis remains largely unresolved. We present a maternal genome-wide meta-analysis of gestational duration (n = 195,555), identifying 22 associated loci (24 independent variants) and an enrichment in genes differentially expressed during labor. A meta-analysis of preterm delivery (18,797 cases, 260,246 controls) revealed six associated loci and large genetic similarities with gestational duration. Analysis of the parental transmitted and nontransmitted alleles (n = 136,833) shows that 15 of the gestational duration genetic variants act through the maternal genome, whereas 7 act both through the maternal and fetal genomes and 2 act only via the fetal genome. Finally, the maternal effects on gestational duration show signs of antagonistic pleiotropy with the fetal effects on birth weight: maternal alleles that increase gestational duration have negative fetal effects on birth weight. The present study provides insights into the genetic effects on the timing of parturition and the complex maternal-fetal relationship between gestational duration and birth weight.
© 2023. The Author(s).
Conflict of interest statement
As of January 2020, A.M. is an employee of Genentech and a holder of Roche stock. The views expressed in this article are those of the author(s) and not necessarily those of the NHS, NIHR or Department of Health. M.I.M. has served on advisory panels for Pfizer, Novo Nordisk and Zoe Global; has received honoraria from Merck, Pfizer, Novo Nordisk and Eli Lilly; and has received research funding from Abbvie, AstraZeneca, Boehringer Ingelheim, Eli Lilly, Janssen, Merck, Novo Nordisk, Pfizer, Roche, Sanofi Aventis, Servier and Takeda. As of June 2019, M.I.M. is an employee of Genentech and a holder of Roche stock. D.A.L. receives support from several national and international government and charitable research funders, as well as from Medtronic and Roche Diagnostics for research unrelated to that presented here. H.S. obtained speaker fees from Ferring Pharmaceuticals, Merck A/S, AstraZeneca and Cook Medical. V.S., G.T., G.S., D.F.G., U.T. and K.S. are employees of deCODE genetics/Amgen. The remaining authors declare no competing interests.
Figures
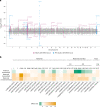
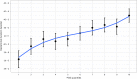
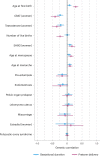

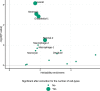

References
Publication types
MeSH terms
Grants and funding
- WT104150/WT_/Wellcome Trust/United Kingdom
- R01 HD056465/HD/NICHD NIH HHS/United States
- MR/P014054/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00011/6/MRC_/Medical Research Council/United Kingdom
- WT104150/WT_/Wellcome Trust/United Kingdom
- R01 AI153356/AI/NIAID NIH HHS/United States
- R01 HD034568/HD/NICHD NIH HHS/United States
- CH/F/20/90003/BHF_/British Heart Foundation/United Kingdom
- AA/18/7/34219/BHF_/British Heart Foundation/United Kingdom
- MC_PC_21038/MRC_/Medical Research Council/United Kingdom
- R01 HD101669/HD/NICHD NIH HHS/United States
- WT220390/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources

