Antigen-specific B cells direct T follicular-like helper cells into lymphoid follicles to mediate Mycobacterium tuberculosis control
- PMID: 37012543
- PMCID: PMC11133959
- DOI: 10.1038/s41590-023-01476-3
Antigen-specific B cells direct T follicular-like helper cells into lymphoid follicles to mediate Mycobacterium tuberculosis control
Abstract
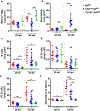
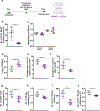
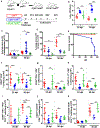
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), is a global cause of death. Granuloma-associated lymphoid tissue (GrALT) correlates with protection during TB, but the mechanisms of protection are not understood. During TB, the transcription factor IRF4 in T cells but not B cells is required for the generation of the TH1 and TH17 subsets of helper T cells and follicular helper T (TFH)-like cellular responses. A population of IRF4+ T cells coexpress the transcription factor BCL6 during Mtb infection, and deletion of Bcl6 (Bcl6fl/fl) in CD4+ T cells (CD4cre) resulted in reduction of TFH-like cells, impaired localization within GrALT and increased Mtb burden. In contrast, the absence of germinal center B cells, MHC class II expression on B cells, antibody-producing plasma cells or interleukin-10-expressing B cells, did not increase Mtb susceptibility. Indeed, antigen-specific B cells enhance cytokine production and strategically localize TFH-like cells within GrALT via interactions between programmed cell death 1 (PD-1) and its ligand PD-L1 and mediate Mtb control in both mice and macaques.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests.
The authors declare no completing interests
Figures















References
-
- WHO. Global Tuberculosis Report (2020).
-
- Scott NR, Swanson RV, Al-Hammadi N, Domingo-Gonzalez R, Rangel-Moreno J, Kriel BA, Bucsan AN, Das S, Ahmed M, Mehra S, Treerat P, Cruz-Lagunas A, Jimenez-Alvarez L, Muñoz-Torrico M, Bobadilla-Lozoya K, Vogl T, Walzl G, du Plessis N, Kaushal D, Scriba TJ, Zúñiga J, Khader SA S100A8/A9 regulates CD11b expression and neutrophil recruitment during chronic tuberculosis. J Clin Invest 130, 3098–3112 (2020). - PMC - PubMed
-
- Slight SR, Rangel-Moreno J, Gopal R, Lin Y, Fallert Junecko BA, Mehra S, Selman M, Becerril-Villanueva E, Baquera-Heredia J, Pavon L, Kaushal D, Reinhart TA, Randall TD, Khader SA CXCR5⁺ T helper cells mediate protective immunity against tuberculosis. J. Clin. Invest. 123, 712–726 (2013). - PMC - PubMed
-
- Esaulova E, Das S, Singh DK, Choreño-Parra JA, Swain A, Arthur L, Rangel-Moreno J, Ahmed M, Singh B, Gupta A, Fernández-López LA, de la Luz Garcia-Hernandez M, Bucsan A, Moodley C, Mehra S, García-Latorre E, Zuniga J, Atkinson J, Kaushal D, Artyomov MN, Khader SA The immune landscape in tuberculosis reveals populations linked to disease and latency. Cell Host Microbe 29, 165–178 e168 (2021). - PMC - PubMed
-
- Ahmed M, Thirunavukkarasu S, Rosa BA, Thomas KA, Das S, Rangel-Moreno J, Lu L, Mehra S, Mbandi SK, Thackray LB, Diamond MS, Murphy KM, Means T, Martin J, Kaushal D, Scriba TJ, Mitreva M, Khader SA Immune correlates of tuberculosis disease and risk translate across species. Sci Transl Med 12, 528 eaay0233 (2020). - PMC - PubMed
Methods References.
-
- Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, Shen F, Eaton SM, Gaffen SL, Swain SL, Locksley RM, Haynes L, Randall TD, Cooper AM. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol 8, 369–377 (2007). - PubMed
-
- Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, Mahomed H, Erasmus M, Whatney W, Hussey GD, Abrahams D, Kafaar F, Hawkridge T, Verver S, Hughes EJ, Ota M, Sutherland J, Howe R, Dockrell HM, Boom WH, Thiel B, Ottenhoff THM, Mayanja-Kizza H, Crampin AC, Downing K, Hatherill M, Valvo J, Shankar S, Parida SK, Kaufmann SHE, Walzl G, Aderem A, Hanekom WA; ACS and GC6–74 cohort study groups. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. The Lancet 387, 2312–2322 (2016). - PMC - PubMed
-
- Mahomed H, Hawkridge T, Verver S, Geiter L, Hatherill M, Abrahams DA, Ehrlich R, Hanekom WA, Hussey GD; SATVI Adolescent Study Team. Predictive factors for latent tuberculosis infection among adolescents in a high-burden area in South Africa. Int J Tuberc Lung Dis 15, 331–336 (2011). - PubMed
-
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, Gil L, Gordon L, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, To JK, Laird MR, Lavidas I, Liu Z, Loveland JE, Maurel T, McLaren W, Moore B, Mudge J, Murphy DN, Newman V, Nuhn M, Ogeh D, Ong CK, Parker A, Patricio M, Riat HS, Schuilenburg H, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Zadissa A, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Cunningham F, Yates A, Flicek P. Ensembl 2018. Nucleic Acids Res 46, D754–D761 (2018). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

