Hypoxia-mediated regulation of DDX5 through decreased chromatin accessibility and post-translational targeting restricts R-loop accumulation
- PMID: 37013907
- PMCID: PMC10323886
- DOI: 10.1002/1878-0261.13431
Hypoxia-mediated regulation of DDX5 through decreased chromatin accessibility and post-translational targeting restricts R-loop accumulation
Abstract
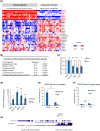
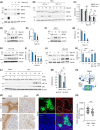
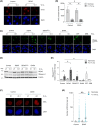
Local hypoxia occurs in most solid tumors and is associated with aggressive disease and therapy resistance. Widespread changes in gene expression play a critical role in the biological response to hypoxia. However, most research has focused on hypoxia-inducible genes as opposed to those that are decreased in hypoxia. We demonstrate that chromatin accessibility is decreased in hypoxia, predominantly at gene promoters and specific pathways are impacted including DNA repair, splicing, and the R-loop interactome. One of the genes with decreased chromatin accessibility in hypoxia was DDX5, encoding the RNA helicase, DDX5, which showed reduced expression in various cancer cell lines in hypoxic conditions, tumor xenografts, and in patient samples with hypoxic tumors. Most interestingly, we found that when DDX5 is rescued in hypoxia, replication stress and R-loop levels accumulate further, demonstrating that hypoxia-mediated repression of DDX5 restricts R-loop accumulation. Together these data support the hypothesis that a critical part of the biological response to hypoxia is the repression of multiple R-loop processing factors; however, as shown for DDX5, their role is specific and distinct.
Keywords: ATACseq; DDX5; R-loops; hypoxia.
© 2023 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

