Translation regulation of specific mRNAs by RPS26 C-terminal RNA-binding tail integrates energy metabolism and AMPK-mTOR signaling
- PMID: 37013984
- PMCID: PMC10201367
- DOI: 10.1093/nar/gkad238
Translation regulation of specific mRNAs by RPS26 C-terminal RNA-binding tail integrates energy metabolism and AMPK-mTOR signaling
Abstract
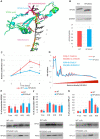
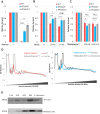
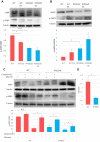
Increasing evidence suggests that ribosome composition and modifications contribute to translation control. Whether direct mRNA binding by ribosomal proteins regulates the translation of specific mRNA and contributes to ribosome specialization has been poorly investigated. Here, we used CRISPR-Cas9 to mutate the RPS26 C-terminus (RPS26dC) predicted to bind AUG upstream nucleotides at the exit channel. RPS26 binding to positions -10 to -16 of short 5' untranslated region (5'UTR) mRNAs exerts positive and negative effects on translation directed by Kozak and Translation Initiator of Short 5'UTR (TISU), respectively. Consistent with that, shortening the 5'UTR from 16 to 10 nt diminished Kozak and enhanced TISU-driven translation. As TISU is resistant and Kozak is sensitive to energy stress, we examined stress responses and found that the RPS26dC mutation confers resistance to glucose starvation and mTOR inhibition. Furthermore, the basal mTOR activity is reduced while AMP-activated protein kinase is activated in RPS26dC cells, mirroring energy-deprived wild-type (WT) cells. Likewise, the translatome of RPS26dC cells is correlated to glucose-starved WT cells. Our findings uncover the central roles of RPS26 C-terminal RNA binding in energy metabolism, in the translation of mRNAs bearing specific features and in the translation tolerance of TISU genes to energy stress.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Hinnebusch A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014; 83:779–812. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

