Proteomic cardiovascular risk assessment in chronic kidney disease
- PMID: 37014015
- PMCID: PMC10281556
- DOI: 10.1093/eurheartj/ehad115
Proteomic cardiovascular risk assessment in chronic kidney disease
Abstract
Aims: Chronic kidney disease (CKD) is widely prevalent and independently increases cardiovascular risk. Cardiovascular risk prediction tools derived in the general population perform poorly in CKD. Through large-scale proteomics discovery, this study aimed to create more accurate cardiovascular risk models.
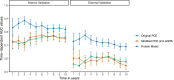
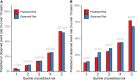
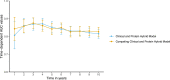
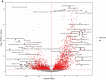
Methods and results: Elastic net regression was used to derive a proteomic risk model for incident cardiovascular risk in 2182 participants from the Chronic Renal Insufficiency Cohort. The model was then validated in 485 participants from the Atherosclerosis Risk in Communities cohort. All participants had CKD and no history of cardiovascular disease at study baseline when ∼5000 proteins were measured. The proteomic risk model, which consisted of 32 proteins, was superior to both the 2013 ACC/AHA Pooled Cohort Equation and a modified Pooled Cohort Equation that included estimated glomerular filtrate rate. The Chronic Renal Insufficiency Cohort internal validation set demonstrated annualized receiver operating characteristic area under the curve values from 1 to 10 years ranging between 0.84 and 0.89 for the protein and 0.70 and 0.73 for the clinical models. Similar findings were observed in the Atherosclerosis Risk in Communities validation cohort. For nearly half of the individual proteins independently associated with cardiovascular risk, Mendelian randomization suggested a causal link to cardiovascular events or risk factors. Pathway analyses revealed enrichment of proteins involved in immunologic function, vascular and neuronal development, and hepatic fibrosis.
Conclusion: In two sizeable populations with CKD, a proteomic risk model for incident cardiovascular disease surpassed clinical risk models recommended in clinical practice, even after including estimated glomerular filtration rate. New biological insights may prioritize the development of therapeutic strategies for cardiovascular risk reduction in the CKD population.
Keywords: Cardiovascular risk; Kidney disease; Mendelian Randomization; Pathway analysis; Prediction; Proteomics.
© The Author(s) 2023. Published by Oxford University Press on behalf of the European Society of Cardiology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest P.G. serves on a medical advisory board to SomaLogic, Inc. for which he accepts no salary, honoraria, or any other financial incentives. J.C. is a scientific advisor to SomaLogic, Inc. S.L.S. serves as a consultant to Agios, FORMA Therapeutics, Global Blood Therapeutics, NOVARTIS, and ORIC Pharmaceuticals.
Figures




Comment in
-
Revolutionizing cardiovascular risk prediction in patients with chronic kidney disease: machine learning and large-scale proteomic risk prediction model lead the way.Eur Heart J. 2023 Jun 20;44(23):2111-2113. doi: 10.1093/eurheartj/ehad127. Eur Heart J. 2023. PMID: 37012642 No abstract available.
References
-
- Grundy SM, Stone NJ, Bailey AL, Beam C, Birtcher KK, Blumenthal RS, et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA guideline on the management of blood cholesterol: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation 2019;139:e1082–e1143. 10.1161/CIR.0000000000000625 - DOI - PMC - PubMed
-
- Whelton PK, Carey RM, Aronow WS, Casey DE Jr, Collins KJ, Dennison Himmelfarb C, et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA guideline for the prevention, detection, evaluation, and management of high blood pressure in adults: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol 2018;71:e127–e248. 10.1016/j.jacc.2017.11.006 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
- R01 HL153161/HL/NHLBI NIH HHS/United States
- U01 DK060963/DK/NIDDK NIH HHS/United States
- UL1 RR024131/RR/NCRR NIH HHS/United States
- U01 DK061022/DK/NIDDK NIH HHS/United States
- R01 DK119199/DK/NIDDK NIH HHS/United States
- UL1 TR000003/TR/NCATS NIH HHS/United States
- UL1 TR000439/TR/NCATS NIH HHS/United States
- UL1 RR029879/RR/NCRR NIH HHS/United States
- U01 DK061028/DK/NIDDK NIH HHS/United States
- UL1 TR000433/TR/NCATS NIH HHS/United States
- U01 DK108809/DK/NIDDK NIH HHS/United States
- UL1 TR002548/TR/NCATS NIH HHS/United States
- U01 DK060984/DK/NIDDK NIH HHS/United States
- U01 DK061021/DK/NIDDK NIH HHS/United States
- U24 DK060990/DK/NIDDK NIH HHS/United States
- U01 DK060980/DK/NIDDK NIH HHS/United States
- R01 HL159081/HL/NHLBI NIH HHS/United States
- U01DK108809 and R01HL159081/NH/NIH HHS/United States
- UL1 TR000424/TR/NCATS NIH HHS/United States
- M01 RR016500/RR/NCRR NIH HHS/United States
- P20 GM109036/GM/NIGMS NIH HHS/United States
- U01 DK060902/DK/NIDDK NIH HHS/United States
- U01 DK060990/DK/NIDDK NIH HHS/United States
- UM1 TR004528/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

