Automatic Discrimination of Species within the Enterobacter cloacae Complex Using Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry and Supervised Algorithms
- PMID: 37014210
- PMCID: PMC10117122
- DOI: 10.1128/jcm.01049-22
Automatic Discrimination of Species within the Enterobacter cloacae Complex Using Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry and Supervised Algorithms
Abstract
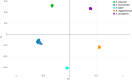
The Enterobacter cloacae complex (ECC) encompasses heterogeneous clusters of species that have been associated with nosocomial outbreaks. These species may have different acquired antimicrobial resistance and virulence mechanisms, and their identification is challenging. This study aims to develop predictive models based on matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) profiles and machine learning for species-level identification. A total of 219 ECC and 118 Klebsiella aerogenes clinical isolates from three hospitals were included. The capability of the proposed method to differentiate the most common ECC species (Enterobacter asburiae, Enterobacter kobei, Enterobacter hormaechei, Enterobacter roggenkampii, Enterobacter ludwigii, and Enterobacter bugandensis) and K. aerogenes was demonstrated by applying unsupervised hierarchical clustering with principal-component analysis (PCA) preprocessing. We observed a distinctive clustering of E. hormaechei and K. aerogenes and a clear trend for the rest of the ECC species to be differentiated over the development data set. Thus, we developed supervised, nonlinear predictive models (support vector machine with radial basis function and random forest). The external validation of these models with protein spectra from two participating hospitals yielded 100% correct species-level assignment for E. asburiae, E. kobei, and E. roggenkampii and between 91.2% and 98.0% for the remaining ECC species; with data analyzed in the three participating centers, the accuracy was close to 100%. Similar results were obtained with the Mass Spectrometric Identification (MSI) database developed recently (https://msi.happy-dev.fr) except in the case of E. hormaechei, which was more accurately identified with the random forest algorithm. In short, MALDI-TOF MS combined with machine learning was demonstrated to be a rapid and accurate method for the differentiation of ECC species.
Keywords: Enterobacter species; MALDI-TOF MS; machine learning; mass spectrometry; peak analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

