CircEPS15, as a sponge of MIR24-3p ameliorates neuronal damage in Parkinson disease through boosting PINK1-PRKN-mediated mitophagy
- PMID: 37014258
- PMCID: PMC10392753
- DOI: 10.1080/15548627.2023.2196889
CircEPS15, as a sponge of MIR24-3p ameliorates neuronal damage in Parkinson disease through boosting PINK1-PRKN-mediated mitophagy
Abstract
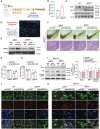
Despite growing evidence that has declared the importance of circRNAs in neurodegenerative diseases, the clinical significance of circRNAs in dopaminergic (DA) neuronal degeneration in the pathogenesis of Parkinson disease (PD) remains unclear. Here, we performed rRNA-depleted RNA sequencing and detected more than 10,000 circRNAs in the plasma samples of PD patients. In consideration of ROC and the correlation between Hohen-Yahr stage (H-Y stage) and Unified Parkinson Disease Rating Scale-motor score (UPDRS) of 40 PD patients, circEPS15 was selected for further research. Low expression of circEPS15 was found in PD patients and there was a negative positive correlation between the circEPS15 level and severity of PD motor symptoms, while overexpression of circEPS15 protected DA neurons against neurotoxin-induced PD-like neurodegeneration in vitro and in vivo. Mechanistically, circEPS15 acted as a MIR24-3p sponge to promote the stable expression of target gene PINK1, thus enhancing PINK1-PRKN-dependent mitophagy to eliminate damaged mitochondria and maintain mitochondrial homeostasis. Thus, circEPS15 rescued DA neuronal degeneration through the MIR24-3p-PINK1 axis-mediated improvement of mitochondrial function. This study reveals that circEPS15 exerts a critical role in participating in PD pathogenesis, and may give us an insight into the novel avenue to develop potential biomarkers and therapeutic targets for PD.Abbreviations: AAV: adeno-associated virus; DA: dopaminergic; FISH: fluorescence in situ hybridizations; HPLC: high-performance liquid chromatography; H-Y stage: Hohen-Yahr stage; LDH: lactate dehydrogenase; MMP: mitochondrial membrane potential; MPTP/p: 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine/probenecid; NC: negative control; PD: Parkinson disease; PINK1: PTEN induced kinase 1; PBS: phosphate-buffered saline; ROS: reactive oxygen species; SNpc: substantia nigra pars compacta; TEM: transmission electron microscopy; UPDRS: Unified Parkinson's Disease Rating Scale-motor score.
Keywords: CircEPS15; MIR24-3p; PINK1; Parkinson disease; mitophagy.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

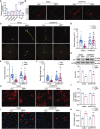
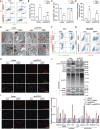

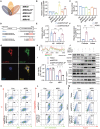
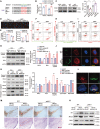


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
