Syrian hamster convalescence from prototype SARS-CoV-2 confers measurable protection against the attenuated disease caused by the Omicron variant
- PMID: 37014911
- PMCID: PMC10104347
- DOI: 10.1371/journal.ppat.1011293
Syrian hamster convalescence from prototype SARS-CoV-2 confers measurable protection against the attenuated disease caused by the Omicron variant
Abstract
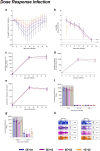
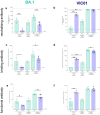
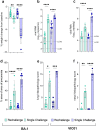
The mutation profile of the SARS-CoV-2 Omicron (lineage BA.1) variant posed a concern for naturally acquired and vaccine-induced immunity. We investigated the ability of prior infection with an early SARS-CoV-2 ancestral isolate (Australia/VIC01/2020, VIC01) to protect against disease caused by BA.1. We established that BA.1 infection in naïve Syrian hamsters resulted in a less severe disease than a comparable dose of the ancestral virus, with fewer clinical signs including less weight loss. We present data to show that these clinical observations were almost absent in convalescent hamsters challenged with the same dose of BA.1 50 days after an initial infection with ancestral virus. These data provide evidence that convalescent immunity against ancestral SARS-CoV-2 is protective against BA.1 in the Syrian hamster model of infection. Comparison with published pre-clinical and clinical data supports consistency of the model and its predictive value for the outcome in humans. Further, the ability to detect protection against the less severe disease caused by BA.1 demonstrates continued value of the Syrian hamster model for evaluation of BA.1-specific countermeasures.
Copyright: © 2023 Ryan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Ryan KA, Bewley KR, Fotheringham SA, Slack GS, Brown P, Hall Y, et al. Dose-dependent response to infection with SARS-CoV-2 in the ferret model and evidence of protective immunity. Nat Commun. 2021;12(1):81. Epub 2021/01/06. doi: 10.1038/s41467-020-20439-y ; PubMed Central PMCID: PMC7782478. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

