Sensitivity optimization of a rhodopsin-based fluorescent voltage indicator
- PMID: 37015225
- PMCID: PMC10280807
- DOI: 10.1016/j.neuron.2023.03.009
Sensitivity optimization of a rhodopsin-based fluorescent voltage indicator
Abstract
The ability to optically image cellular transmembrane voltages at millisecond-timescale resolutions can offer unprecedented insight into the function of living brains in behaving animals. Here, we present a point mutation that increases the sensitivity of Ace2 opsin-based voltage indicators. We use the mutation to develop Voltron2, an improved chemigeneic voltage indicator that has a 65% higher sensitivity to single APs and 3-fold higher sensitivity to subthreshold potentials than Voltron. Voltron2 retained the sub-millisecond kinetics and photostability of its predecessor, although with lower baseline fluorescence. In multiple in vitro and in vivo comparisons with its predecessor across multiple species, we found Voltron2 to be more sensitive to APs and subthreshold fluctuations. Finally, we used Voltron2 to study and evaluate the possible mechanisms of interneuron synchronization in the mouse hippocampus. Overall, we have discovered a generalizable mutation that significantly increases the sensitivity of Ace2 rhodopsin-based sensors, improving their voltage reporting capability.
Keywords: biosensors; fluorescence imaging; fluorescent proteins; genetically encoded indicators; voltage imaging.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.S.A., L.D.L., and E.R.S. have filed for a patent on the chemigenetic voltage indicators. I.K. and C.R.F. are co-inventors on a patent describing pipette cleaning that is licensed by Sensapex. M.C. and L.C. have performed consulting services for Sensapex.
Figures


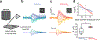

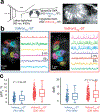


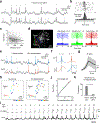
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

