An epigenetic switch controls an alternative NR2F2 isoform that unleashes a metastatic program in melanoma
- PMID: 37015919
- PMCID: PMC10073109
- DOI: 10.1038/s41467-023-36967-2
An epigenetic switch controls an alternative NR2F2 isoform that unleashes a metastatic program in melanoma
Abstract
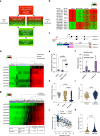
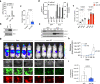
Metastatic melanoma develops once transformed melanocytic cells begin to de-differentiate into migratory and invasive melanoma cells with neural crest cell (NCC)-like and epithelial-to-mesenchymal transition (EMT)-like features. However, it is still unclear how transformed melanocytes assume a metastatic melanoma cell state. Here, we define DNA methylation changes that accompany metastatic progression in melanoma patients and discover Nuclear Receptor Subfamily 2 Group F, Member 2 - isoform 2 (NR2F2-Iso2) as an epigenetically regulated metastasis driver. NR2F2-Iso2 is transcribed from an alternative transcriptional start site (TSS) and it is truncated at the N-terminal end which encodes the NR2F2 DNA-binding domain. We find that NR2F2-Iso2 expression is turned off by DNA methylation when NCCs differentiate into melanocytes. Conversely, this process is reversed during metastatic melanoma progression, when NR2F2-Iso2 becomes increasingly hypomethylated and re-expressed. Our functional and molecular studies suggest that NR2F2-Iso2 drives metastatic melanoma progression by modulating the activity of full-length NR2F2 (Isoform 1) over EMT- and NCC-associated target genes. Our findings indicate that DNA methylation changes play a crucial role during metastatic melanoma progression, and their control of NR2F2 activity allows transformed melanocytes to acquire NCC-like and EMT-like features. This epigenetically regulated transcriptional plasticity facilitates cell state transitions and metastatic spread.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Kath R, Jambrosic JA, Holland L, Rodeck U, Herlyn M. Development of invasive and growth factor-independent cell variants from primary human melanomas. Cancer Res. 1991;51:2205–2211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

