tRF-Gln-CTG-026 ameliorates liver injury by alleviating global protein synthesis
- PMID: 37015921
- PMCID: PMC10073094
- DOI: 10.1038/s41392-023-01351-5
tRF-Gln-CTG-026 ameliorates liver injury by alleviating global protein synthesis
Abstract
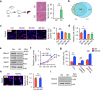
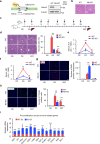
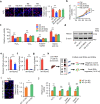
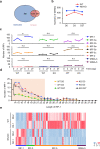
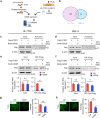
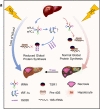
tsRNAs (tRNA-derived small RNAs), as products of the stress response, exert considerable influence on stress response and injury regulation. However, it remains largely unclear whether tsRNAs can ameliorate liver injury. Here, we demonstrate the roles of tsRNAs in alleviating liver injury by utilizing the loss of NSun2 (NOP2/Sun domain family, member 2) as a tsRNAs-generating model. Mechanistically, the loss of NSun2 reduces methyluridine-U5 (m5U) and cytosine-C5 (m5C) of tRNAs, followed by the production of various tsRNAs, especially Class I tsRNAs (tRF-1s). Through further screening, we show that tRF-Gln-CTG-026 (tG026), the optimal tRF-1, ameliorates liver injury by repressing global protein synthesis through the weakened association between TSR1 (pre-rRNA-processing protein TSR1 homolog) and pre-40S ribosome. This study indicates the potential of tsRNA-reduced global protein synthesis in liver injury and repair, suggesting a potential therapeutic strategy for liver injury.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

