Shine: A novel strategy to extract specific, sensitive and well-conserved biomarkers from massive microbial genomic datasets
- PMID: 37016282
- PMCID: PMC10071469
- DOI: 10.1186/s12859-023-05195-2
Shine: A novel strategy to extract specific, sensitive and well-conserved biomarkers from massive microbial genomic datasets
Abstract
Background: Concentrations of the pathogenic microorganisms' DNA in biological samples are typically low. Therefore, DNA diagnostics of common infections are costly, rarely accurate, and challenging. Limited by failing to cover updated epidemic testing samples, computational services are difficult to implement in clinical applications without complex customized settings. Furthermore, the combined biomarkers used to maintain high conservation may not be cost effective and could cause several experimental errors in many clinical settings. Given the limitations of recent developed technology, 16S rRNA is too conserved to distinguish closely related species, and mosaic plasmids are not effective as well because of their uneven distribution across prokaryotic taxa.
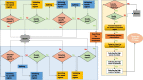
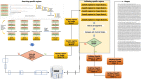
Results: Here, we provide a computational strategy, Shine, that allows extraction of specific, sensitive and well-conserved biomarkers from massive microbial genomic datasets. Distinguished with simple concatenations with blast-based filtering, our method involves a de novo genome alignment-based pipeline to explore the original and specific repetitive biomarkers in the defined population. It can cover all members to detect newly discovered multicopy conserved species-specific or even subspecies-specific target probes and primer sets. The method has been successfully applied to a number of clinical projects and has the overwhelming advantages of automated detection of all pathogenic microorganisms without the limitations of genome annotation and incompletely assembled motifs. Using on our pipeline, users may select different configuration parameters depending on the purpose of the project for routine clinical detection practices on the website https://bioinfo.liferiver.com.cn with easy registration.
Conclusions: The proposed strategy is suitable for identifying shared phylogenetic markers while featuring low rates of false positive or false negative. This technology is suitable for the automatic design of minimal and efficient PCR primers and other types of detection probes.
Keywords: Biomarker; Conservation; Microbial genome; Sensitivity; Specificity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
ezTree: an automated pipeline for identifying phylogenetic marker genes and inferring evolutionary relationships among uncultivated prokaryotic draft genomes.BMC Genomics. 2018 Jan 19;19(Suppl 1):921. doi: 10.1186/s12864-017-4327-9. BMC Genomics. 2018. PMID: 29363425 Free PMC article.
-
[A broad-range 16S rRNA gene real-time PCR assay for the diagnosis of neonatal septicemia].Zhonghua Er Ke Za Zhi. 2007 Jun;45(6):446-9. Zhonghua Er Ke Za Zhi. 2007. PMID: 17880793 Chinese.
-
Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing.mSphere. 2021 Feb 24;6(1):e01202-20. doi: 10.1128/mSphere.01202-20. mSphere. 2021. PMID: 33627512 Free PMC article.
-
Impact of genomics on the understanding of microbial evolution and classification: the importance of Darwin's views on classification.FEMS Microbiol Rev. 2016 Jul;40(4):520-53. doi: 10.1093/femsre/fuw011. Epub 2016 Jun 8. FEMS Microbiol Rev. 2016. PMID: 27279642 Review.
-
Application of Comparative Genomics for the Development of PCR Primers for the Detection of Harmful or Beneficial Microorganisms in Food: Mini-Review.Foods. 2025 Mar 20;14(6):1060. doi: 10.3390/foods14061060. Foods. 2025. PMID: 40232097 Free PMC article. Review.
References
-
- Fox RTV. The present and future use of technology to detect plant pathogens to guide disease control in sustainable farming systems. Agr Ecosyst Environ. 1997;64(2):125–132.
-
- Anahtar MN, Shaw BM, Slater D, Byrne EH, Botti-Lodovico Y, Adams G, et al. Development of a qualitative real-time RT-PCR assay for the detection of SARS-CoV-2: a guide and case study in setting up an emergency-use, laboratory-developed molecular microbiological assay. Journal of Clinical Pathology, 2021: p. jclinpath-2020-207128. - PMC - PubMed
-
- Rajapaksha P, et al. A review of methods for the detection of pathogenic microorganisms. Analyst. 2019;144(2):396–411. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

