Nuclear genetic background influences the phenotype of the Drosophila tko25t mitochondrial protein-synthesis mutant
- PMID: 37017029
- PMCID: PMC10234395
- DOI: 10.1093/g3journal/jkad078
Nuclear genetic background influences the phenotype of the Drosophila tko25t mitochondrial protein-synthesis mutant
Abstract
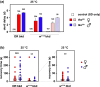
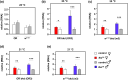
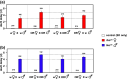
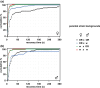
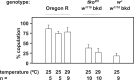
The Drosophila tko25t point mutation in the gene encoding mitoribosomal protein S12 produces a complex phenotype of multiple respiratory chain deficiency, developmental delay, bang-sensitivity, impaired hearing, sugar and antibiotic sensitivity, and impaired male courtship. Its phenotypic severity was previously shown to be alleviated by inbreeding and to vary with mitochondrial genetic background. Here, we show similarly profound effects conferred by nuclear genetic background. We backcrossed tko25t into each of 2 standard nuclear backgrounds, Oregon R and w1118, the latter used as recipient line in many transgenic applications requiring selection for the white minigene marker. In the w1118 background, tko25t flies showed a moderate developmental delay and modest bang-sensitivity. In the Oregon R background, males showed longer developmental delay and more severe bang-sensitivity, and we were initially unable to produce homozygous tko25t females in sufficient numbers to conduct a meaningful analysis. When maintained as a balanced stock over 2 years, tko25t flies in the Oregon R background showed clear phenotypic improvement though were still more severely affected than in the w1118 background. Phenotypic severity did not correlate with the expression level of the tko gene. Analysis of tko25t hybrids between the 2 backgrounds indicated that phenotypic severity was conferred by autosomal, X-chromosomal, and parent-of-origin-dependent determinants. Although some of these effects may be tko25t specific, we recommend that, in order to minimize genetic drift and confounding background effects, the genetic background of nonlethal mutants should be controlled by regular backcrossing, even if stocks are usually maintained over a balancer chromosome.
Keywords: mitochondria; nuclear background; ribosome; semilethality.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflict of interest.
Figures


Similar articles
-
The alternative oxidase AOX does not rescue the phenotype of tko25t mutant flies.G3 (Bethesda). 2014 Aug 21;4(10):2013-21. doi: 10.1534/g3.114.013946. G3 (Bethesda). 2014. PMID: 25147191 Free PMC article.
-
Technical knockout, a Drosophila model of mitochondrial deafness.Genetics. 2001 Sep;159(1):241-54. doi: 10.1093/genetics/159.1.241. Genetics. 2001. PMID: 11560901 Free PMC article.
-
Phenotypic suppression of the Drosophila mitochondrial disease-like mutant tko(25t) by duplication of the mutant gene in its natural chromosomal context.Mitochondrion. 2009 Sep;9(5):353-63. doi: 10.1016/j.mito.2009.07.002. Epub 2009 Jul 17. Mitochondrion. 2009. PMID: 19616644
-
A cytoplasmic suppressor of a nuclear mutation affecting mitochondrial functions in Drosophila.Genetics. 2012 Oct;192(2):483-93. doi: 10.1534/genetics.112.143719. Epub 2012 Jul 30. Genetics. 2012. PMID: 22851652 Free PMC article.
-
Mitochondrial disease in flies.Biochim Biophys Acta. 2004 Dec 6;1659(2-3):190-6. doi: 10.1016/j.bbabio.2004.07.004. Biochim Biophys Acta. 2004. PMID: 15576051 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
