mRNA Regulation by RNA Modifications
- PMID: 37018844
- PMCID: PMC11192554
- DOI: 10.1146/annurev-biochem-052521-035949
mRNA Regulation by RNA Modifications
Abstract
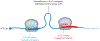
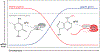
Chemical modifications on mRNA represent a critical layer of gene expression regulation. Research in this area has continued to accelerate over the last decade, as more modifications are being characterized with increasing depth and breadth. mRNA modifications have been demonstrated to influence nearly every step from the early phases of transcript synthesis in the nucleus through to their decay in the cytoplasm, but in many cases, the molecular mechanisms involved in these processes remain mysterious. Here, we highlight recent work that has elucidated the roles of mRNA modifications throughout the mRNA life cycle, describe gaps in our understanding and remaining open questions, and offer some forward-looking perspective on future directions in the field.
Keywords: RNA metabolism; RNA modifications; epitranscriptome; posttranscriptional regulation.
Figures

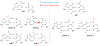

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

