STING mediates hepatocyte pyroptosis in liver fibrosis by Epigenetically activating the NLRP3 inflammasome
- PMID: 37018971
- PMCID: PMC10106968
- DOI: 10.1016/j.redox.2023.102691
STING mediates hepatocyte pyroptosis in liver fibrosis by Epigenetically activating the NLRP3 inflammasome
Abstract
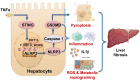
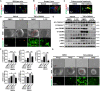
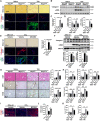
The activation of stimulator of interferon genes (STING) and NOD-like receptor protein 3 (NLRP3) inflammasome-mediated pyroptosis signaling pathways represent two distinct central mechanisms in liver disease. However, the interconnections between these two pathways and the epigenetic regulation of the STING-NLRP3 axis in hepatocyte pyroptosis during liver fibrosis remain unknown. STING and NLRP3 inflammasome signaling pathways are activated in fibrotic livers but are suppressed by Sting knockout. Sting knockout ameliorated hepatic pyroptosis, inflammation, and fibrosis. In vitro, STING induces pyroptosis in primary murine hepatocytes by activating the NLRP3 inflammasome. H3K4-specific histone methyltransferase WD repeat-containing protein 5 (WDR5) and DOT1-like histone H3K79 methyltransferase (DOT1L) are identified to regulate NLRP3 expression in STING-overexpressing AML12 hepatocytes. WDR5/DOT1L-mediated histone methylation enhances interferon regulatory transcription factor 3 (IRF3) binding to the Nlrp3 promoter and promotes STING-induced Nlrp3 transcription in hepatocytes. Moreover, hepatocyte-specific Nlrp3 deletion and downstream Gasdermin D (Gsdmd) knockout attenuate hepatic pyroptosis, inflammation, and fibrosis. RNA-sequencing and metabolomics analysis in murine livers and primary hepatocytes show that oxidative stress and metabolic reprogramming might participate in NLRP3-mediated hepatocyte pyroptosis and liver fibrosis. The STING-NLRP3-GSDMD axis inhibition suppresses hepatic ROS generation. In conclusion, this study describes a novel epigenetic mechanism by which the STING-WDR5/DOT1L/IRF3-NLRP3 signaling pathway enhances hepatocyte pyroptosis and hepatic inflammation in liver fibrosis.
Keywords: GSDMD; Histone methylation; IRF3; Liver cirrhosis; Metabolic reprogramming; Oxidative stress.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest All authors declare no conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

