Generation of ventralized human thalamic organoids with thalamic reticular nucleus
- PMID: 37019105
- PMCID: PMC10329908
- DOI: 10.1016/j.stem.2023.03.007
Generation of ventralized human thalamic organoids with thalamic reticular nucleus
Abstract
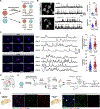
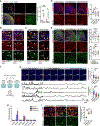
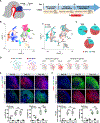
Human brain organoids provide unique platforms for modeling several aspects of human brain development and pathology. However, current brain organoid systems mostly lack the resolution to recapitulate the development of finer brain structures with subregional identity, including functionally distinct nuclei in the thalamus. Here, we report a method for converting human embryonic stem cells (hESCs) into ventral thalamic organoids (vThOs) with transcriptionally diverse nuclei identities. Notably, single-cell RNA sequencing revealed previously unachieved thalamic patterning with a thalamic reticular nucleus (TRN) signature, a GABAergic nucleus located in the ventral thalamus. Using vThOs, we explored the functions of TRN-specific, disease-associated genes patched domain containing 1 (PTCHD1) and receptor tyrosine-protein kinase (ERBB4) during human thalamic development. Perturbations in PTCHD1 or ERBB4 impaired neuronal functions in vThOs, albeit not affecting the overall thalamic lineage development. Together, vThOs present an experimental model for understanding nuclei-specific development and pathology in the thalamus of the human brain.
Keywords: 3-D organoids; ERBB4; PTCHD1; TRN; brain organoid; hESC; stem cells; thalamic organoid; thalamic reticular nucleus; ventral thalamic organoids.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

Similar articles
-
hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids.Cell Stem Cell. 2019 Mar 7;24(3):487-497.e7. doi: 10.1016/j.stem.2018.12.015. Epub 2019 Feb 21. Cell Stem Cell. 2019. PMID: 30799279 Free PMC article.
-
Activating PV-positive neurons in ventral thalamic reticular nucleus reduces pain sensitivity in mice.Brain Res. 2023 Jan 15;1799:148174. doi: 10.1016/j.brainres.2022.148174. Epub 2022 Nov 24. Brain Res. 2023. PMID: 36427592
-
Organization in the somatosensory sector of the cat's thalamic reticular nucleus.J Comp Neurol. 1996 Mar 4;366(2):207-22. doi: 10.1002/(SICI)1096-9861(19960304)366:2<207::AID-CNE2>3.0.CO;2-9. J Comp Neurol. 1996. PMID: 8698882
-
Thalamic reticular nucleus in the thalamocortical loop.Neurosci Res. 2020 Jul;156:32-40. doi: 10.1016/j.neures.2019.12.004. Epub 2019 Dec 5. Neurosci Res. 2020. PMID: 31812650 Review.
-
The thalamic reticular nucleus and schizophrenia.Schizophr Bull. 2011 Mar;37(2):306-15. doi: 10.1093/schbul/sbq142. Epub 2010 Dec 3. Schizophr Bull. 2011. PMID: 21131368 Free PMC article. Review.
Cited by
-
Human Brain Organoids: Development and Applications.J Microbiol Biotechnol. 2025 May 28;35:e2411040. doi: 10.4014/jmb.2411.11040. J Microbiol Biotechnol. 2025. PMID: 40443221 Free PMC article. Review.
-
Multiscale engineering of brain organoids for disease modeling.Adv Drug Deliv Rev. 2024 Jul;210:115344. doi: 10.1016/j.addr.2024.115344. Epub 2024 May 27. Adv Drug Deliv Rev. 2024. PMID: 38810702 Free PMC article. Review.
-
Midbrain organoids-development and applications in Parkinson's disease.Oxf Open Neurosci. 2023 Aug 18;2:kvad009. doi: 10.1093/oons/kvad009. eCollection 2023. Oxf Open Neurosci. 2023. PMID: 38596240 Free PMC article. Review.
-
Exploring human brain development and disease using assembloids.Neuron. 2025 Apr 16;113(8):1133-1150. doi: 10.1016/j.neuron.2025.02.010. Epub 2025 Mar 18. Neuron. 2025. PMID: 40107269 Review.
-
The promise of cerebral organoids for neonatology.Curr Opin Pediatr. 2025 Apr 1;37(2):182-190. doi: 10.1097/MOP.0000000000001446. Epub 2025 Feb 12. Curr Opin Pediatr. 2025. PMID: 40013913 Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

