Targeting PPAR-gamma counteracts tumour adaptation to immune-checkpoint blockade in hepatocellular carcinoma
- PMID: 37019619
- PMCID: PMC10423534
- DOI: 10.1136/gutjnl-2022-328364
Targeting PPAR-gamma counteracts tumour adaptation to immune-checkpoint blockade in hepatocellular carcinoma
Abstract
Objective: Therapy-induced tumour microenvironment (TME) remodelling poses a major hurdle for cancer cure. As the majority of patients with hepatocellular carcinoma (HCC) exhibits primary or acquired resistance to antiprogrammed cell death (ligand)-1 (anti-PD-[L]1) therapies, we aimed to investigate the mechanisms underlying tumour adaptation to immune-checkpoint targeting.
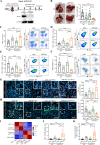
Design: Two immunotherapy-resistant HCC models were generated by serial orthotopic implantation of HCC cells through anti-PD-L1-treated syngeneic, immunocompetent mice and interrogated by single-cell RNA sequencing (scRNA-seq), genomic and immune profiling. Key signalling pathway was investigated by lentiviral-mediated knockdown and pharmacological inhibition, and further verified by scRNA-seq analysis of HCC tumour biopsies from a phase II trial of pembrolizumab (NCT03419481).
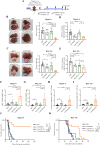
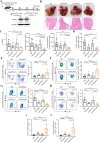
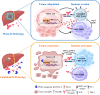
Results: Anti-PD-L1-resistant tumours grew >10-fold larger than parental tumours in immunocompetent but not immunocompromised mice without overt genetic changes, which were accompanied by intratumoral accumulation of myeloid-derived suppressor cells (MDSC), cytotoxic to exhausted CD8+ T cell conversion and exclusion. Mechanistically, tumour cell-intrinsic upregulation of peroxisome proliferator-activated receptor-gamma (PPARγ) transcriptionally activated vascular endothelial growth factor-A (VEGF-A) production to drive MDSC expansion and CD8+ T cell dysfunction. A selective PPARγ antagonist triggered an immune suppressive-to-stimulatory TME conversion and resensitised tumours to anti-PD-L1 therapy in orthotopic and spontaneous HCC models. Importantly, 40% (6/15) of patients with HCC resistant to pembrolizumab exhibited tumorous PPARγ induction. Moreover, higher baseline PPARγ expression was associated with poorer survival of anti-PD-(L)1-treated patients in multiple cancer types.
Conclusion: We uncover an adaptive transcriptional programme by which tumour cells evade immune-checkpoint targeting via PPARγ/VEGF-A-mediated TME immunosuppression, thus providing a strategy for counteracting immunotherapeutic resistance in HCC.
Keywords: CANCER IMMUNOBIOLOGY; HEPATOCELLULAR CARCINOMA; IMMUNOTHERAPY; PPAR GAMMA.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: The authors declare no conflicts of interest that pertain to this work except the following declarations. YMDL is a scientific cofounder of Grail, receives royalties from Illumina, Grail, Sequenom, Xcelom, DRA and Take2, serves as a consultant to Decheng Capital and holds equity in Illumina/Grail, DRA and Take2. SLC serves as a consultant to and receives honoraria from Astra-Zeneca, Eisai and MSD.
Figures




References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
